Detail Information of Epigenetic Regulations
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0531 Transporter Info | ||||
| Gene Name | KCNH7 | ||||
| Transporter Name | Voltage-gated potassium channel Kv11.3 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| Epigenetic Regulations of This DT (EGR) | |||||
|---|---|---|---|---|---|
|
Methylation |
|||||
|
Bladder cancer |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of KCNH7 in bladder cancer | [ 1 ] | |||
|
Location |
Body (cg23626131) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.03E+00 | Statistic Test | p-value: 2.56E-02; Z-score: -7.18E-01 | ||
|
Methylation in Case |
7.50E-01 (Median) | Methylation in Control | 7.73E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of KCNH7 in bladder cancer | [ 1 ] | |||
|
Location |
3'UTR (cg14983936) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.19E+00 | Statistic Test | p-value: 1.36E-03; Z-score: -3.51E+00 | ||
|
Methylation in Case |
6.77E-01 (Median) | Methylation in Control | 8.04E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Breast cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of KCNH7 in breast cancer | [ 2 ] | |||
|
Location |
Body (cg23626131) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.06E+00 | Statistic Test | p-value: 1.07E-02; Z-score: -5.71E-01 | ||
|
Methylation in Case |
7.44E-01 (Median) | Methylation in Control | 7.87E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colorectal cancer |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of KCNH7 in colorectal cancer | [ 3 ] | |||
|
Location |
Body (cg23626131) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.11E+00 | Statistic Test | p-value: 1.29E-08; Z-score: -3.28E+00 | ||
|
Methylation in Case |
8.02E-01 (Median) | Methylation in Control | 8.88E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of KCNH7 in colorectal cancer | [ 3 ] | |||
|
Location |
3'UTR (cg14983936) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.07E+00 | Statistic Test | p-value: 2.57E-07; Z-score: -2.15E+00 | ||
|
Methylation in Case |
8.24E-01 (Median) | Methylation in Control | 8.79E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Hepatocellular carcinoma |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of KCNH7 in hepatocellular carcinoma | [ 4 ] | |||
|
Location |
Body (cg14914532) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.31E+00 | Statistic Test | p-value: 1.57E-12; Z-score: -4.95E+00 | ||
|
Methylation in Case |
5.79E-01 (Median) | Methylation in Control | 7.59E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of KCNH7 in hepatocellular carcinoma | [ 4 ] | |||
|
Location |
3'UTR (cg14983936) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.16E+00 | Statistic Test | p-value: 1.73E-09; Z-score: -2.05E+00 | ||
|
Methylation in Case |
6.76E-01 (Median) | Methylation in Control | 7.86E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Pancretic ductal adenocarcinoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of KCNH7 in pancretic ductal adenocarcinoma | [ 5 ] | |||
|
Location |
Body (cg05073386) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.01E+00 | Statistic Test | p-value: 2.26E-03; Z-score: -6.27E-01 | ||
|
Methylation in Case |
8.64E-01 (Median) | Methylation in Control | 8.73E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Lung adenocarcinoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of KCNH7 in lung adenocarcinoma | [ 6 ] | |||
|
Location |
3'UTR (cg14983936) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.03E+00 | Statistic Test | p-value: 2.25E-02; Z-score: -8.07E-01 | ||
|
Methylation in Case |
8.17E-01 (Median) | Methylation in Control | 8.39E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Papillary thyroid cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of KCNH7 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
3'UTR (cg14983936) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.04E+00 | Statistic Test | p-value: 3.82E-02; Z-score: 9.29E-01 | ||
|
Methylation in Case |
8.77E-01 (Median) | Methylation in Control | 8.47E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Third ventricle chordoid glioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
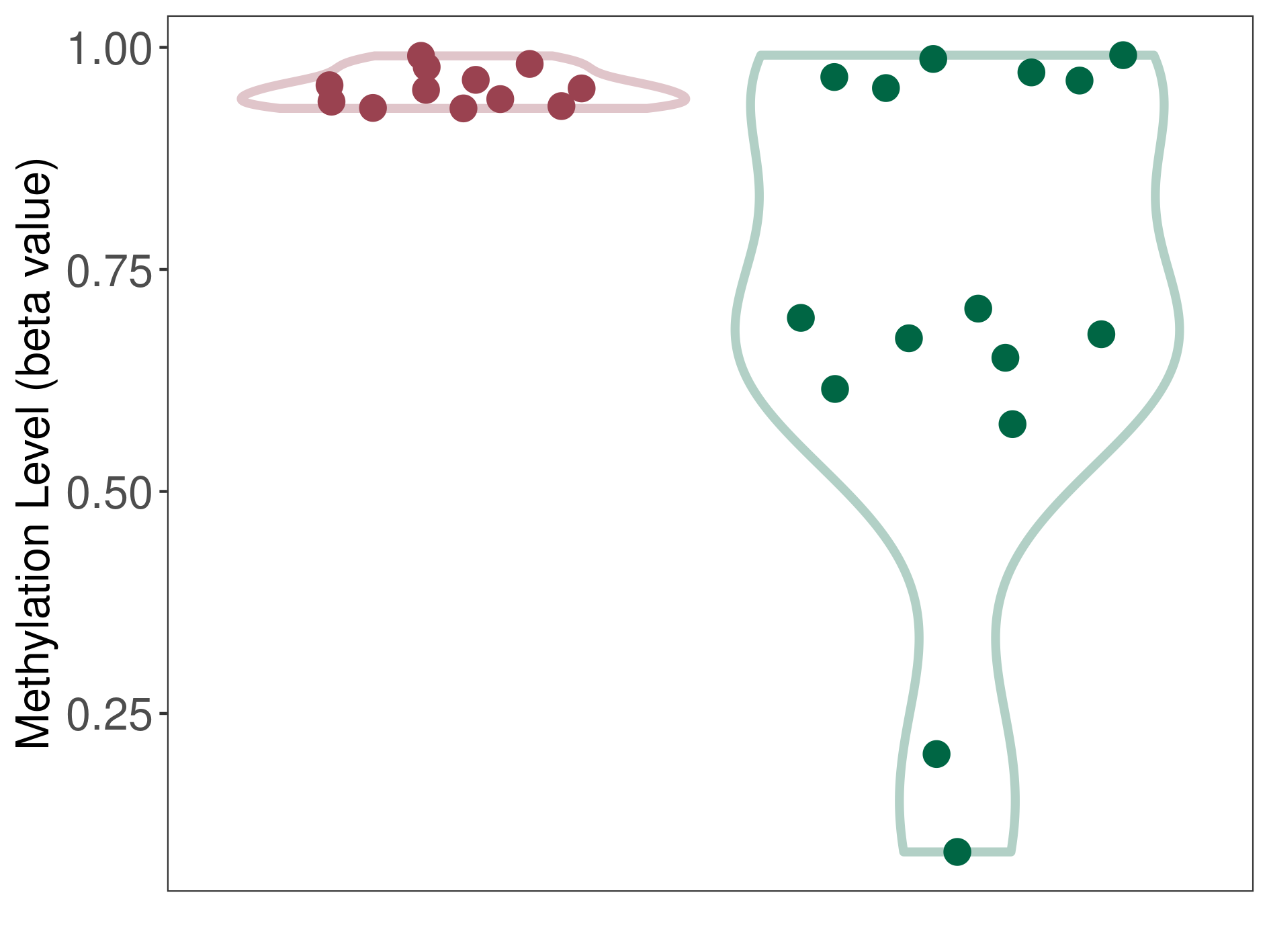
Epigenetic Phenomenon 1 |
Moderate hypermethylation of KCNH7 in third ventricle chordoid glioma than that in healthy individual | ||||
Studied Phenotype |
Third ventricle chordoid glioma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 0.004908653; Fold-change: 0.257505468; Z-score: 0.926471889 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
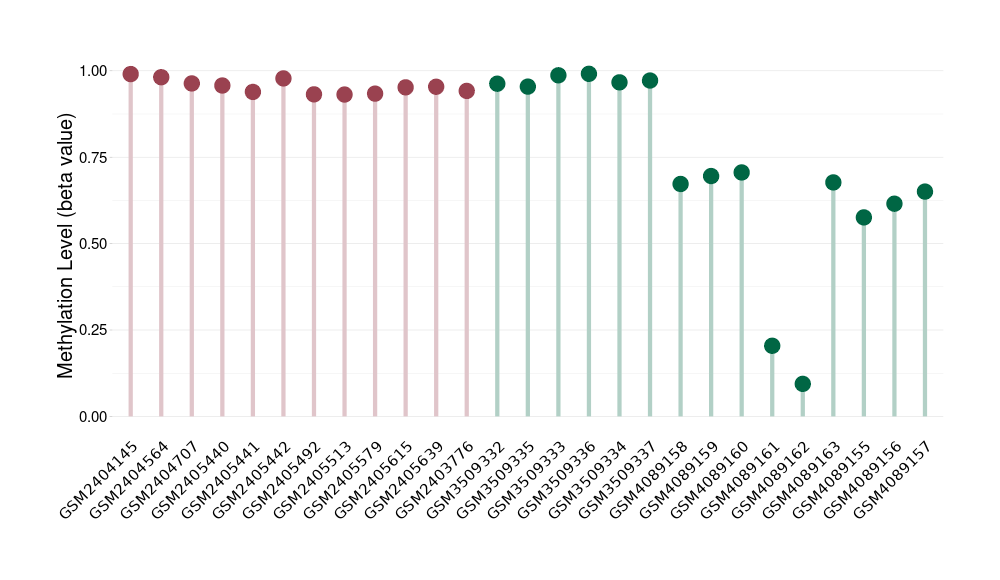 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Lymphoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
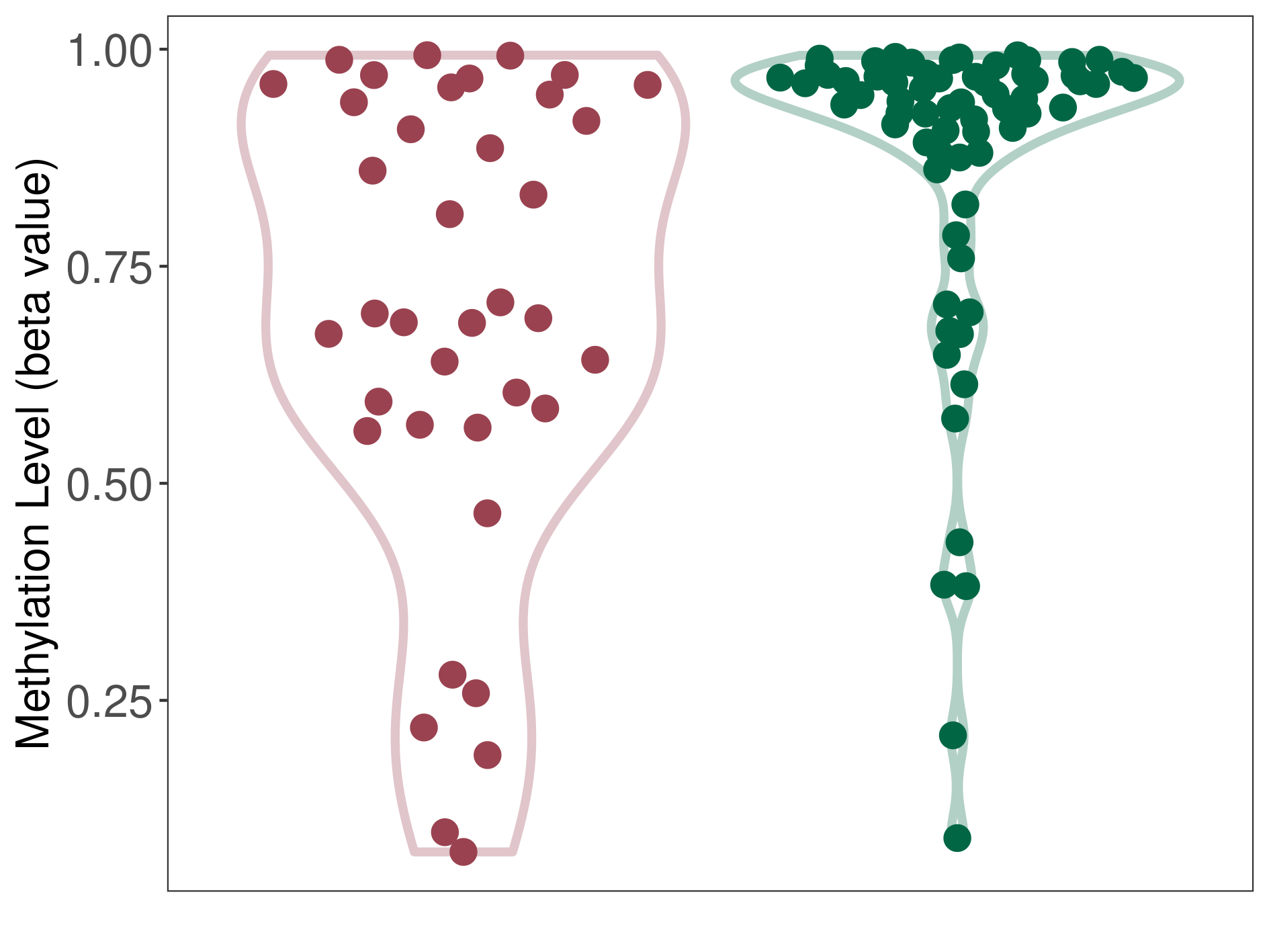
Epigenetic Phenomenon 1 |
Moderate hypomethylation of KCNH7 in lymphoma than that in healthy individual | ||||
Studied Phenotype |
Lymphoma [ICD-11:2B30] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 0.001198133; Fold-change: -0.247099715; Z-score: -1.280925928 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
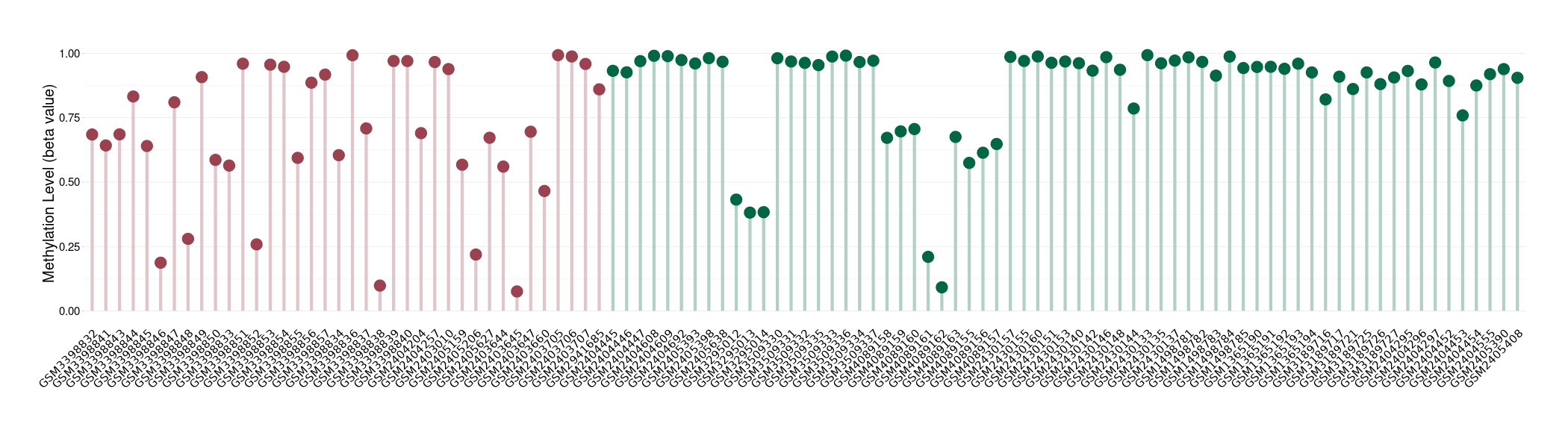 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
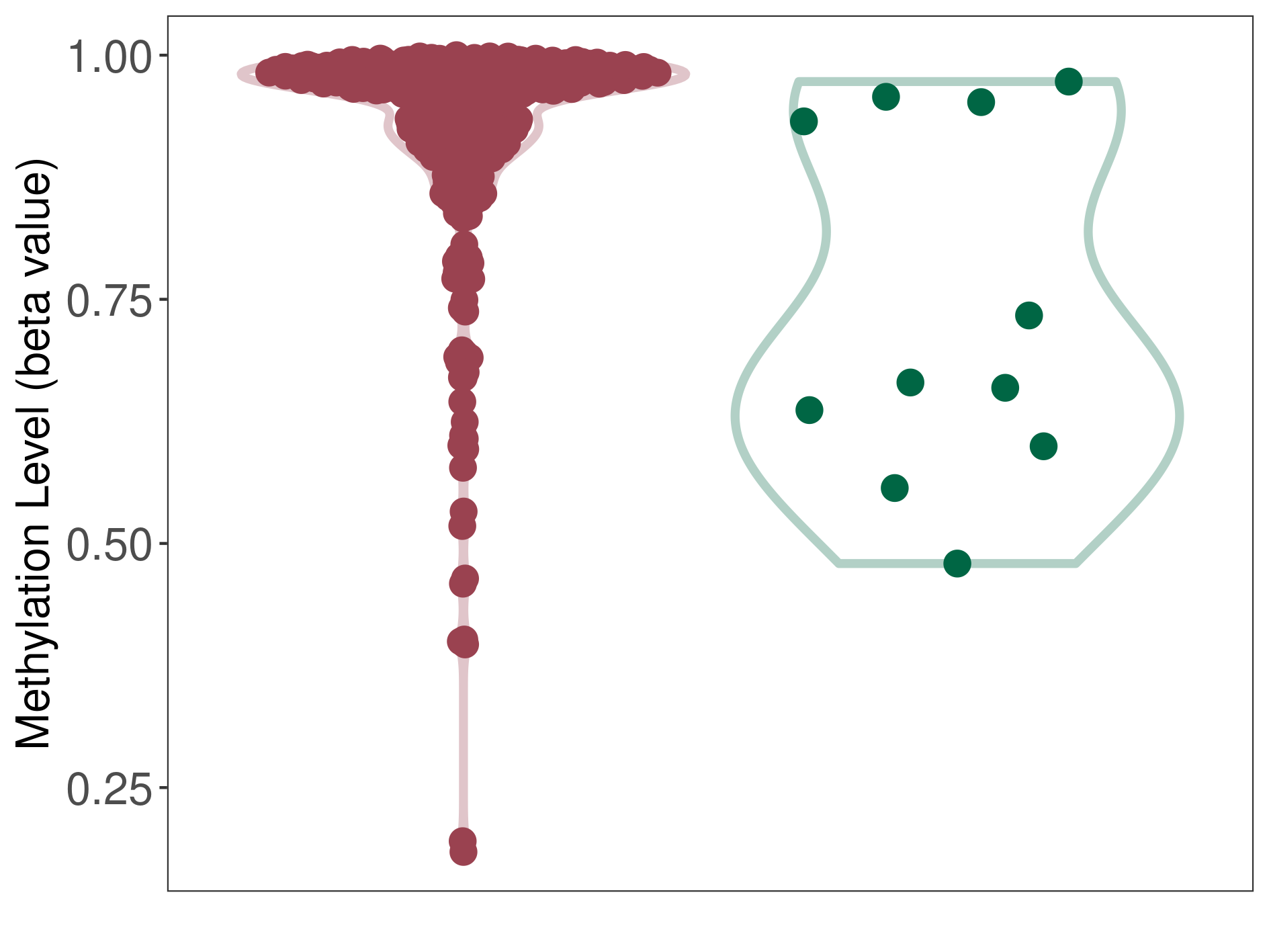
Prostate cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypermethylation of KCNH7 in prostate cancer than that in healthy individual | ||||
Studied Phenotype |
Prostate cancer [ICD-11:2C82] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 0.006429563; Fold-change: 0.301374926; Z-score: 1.665310147 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
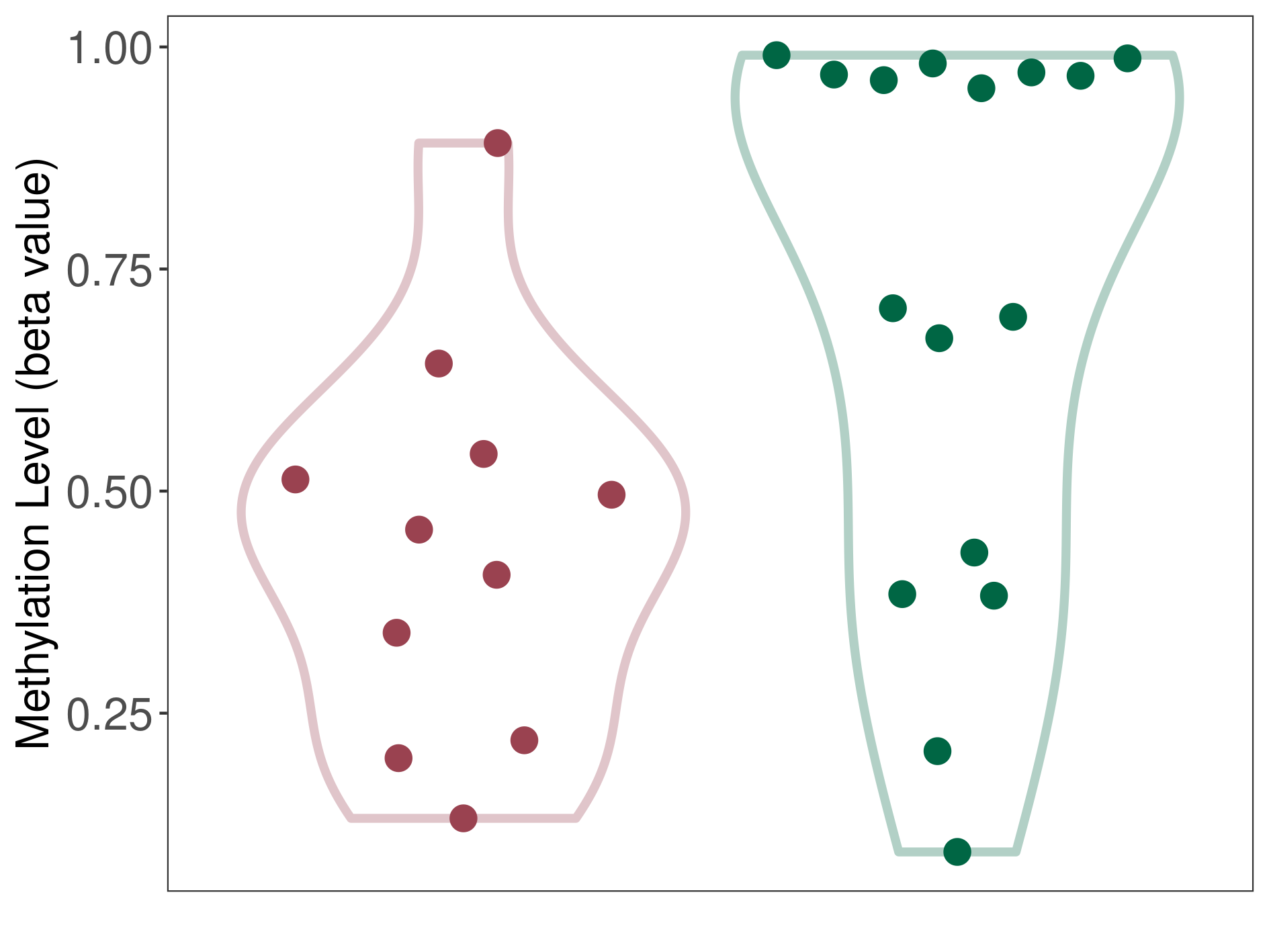
Cerebellar liponeurocytoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypomethylation of KCNH7 in cerebellar liponeurocytoma than that in healthy individual | ||||
Studied Phenotype |
Cerebellar liponeurocytoma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 0.01434866; Fold-change: -0.373056701; Z-score: -1.187739255 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
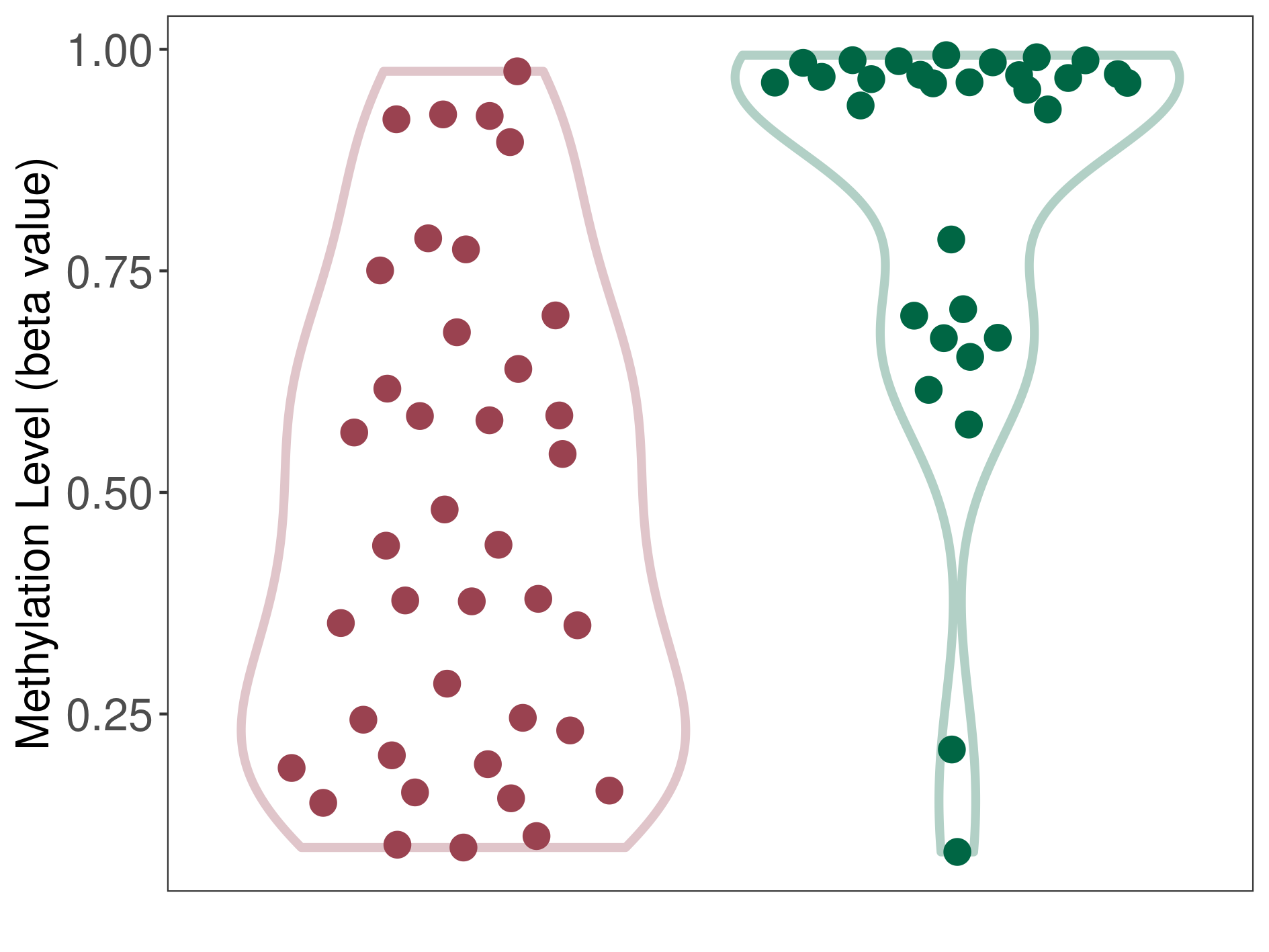
Esthesioneuroblastoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypomethylation of KCNH7 in esthesioneuroblastoma than that in healthy individual | ||||
Studied Phenotype |
Esthesioneuroblastoma [ICD-11:2D50.1] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 5.81E-08; Fold-change: -0.522381882; Z-score: -2.26130328 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
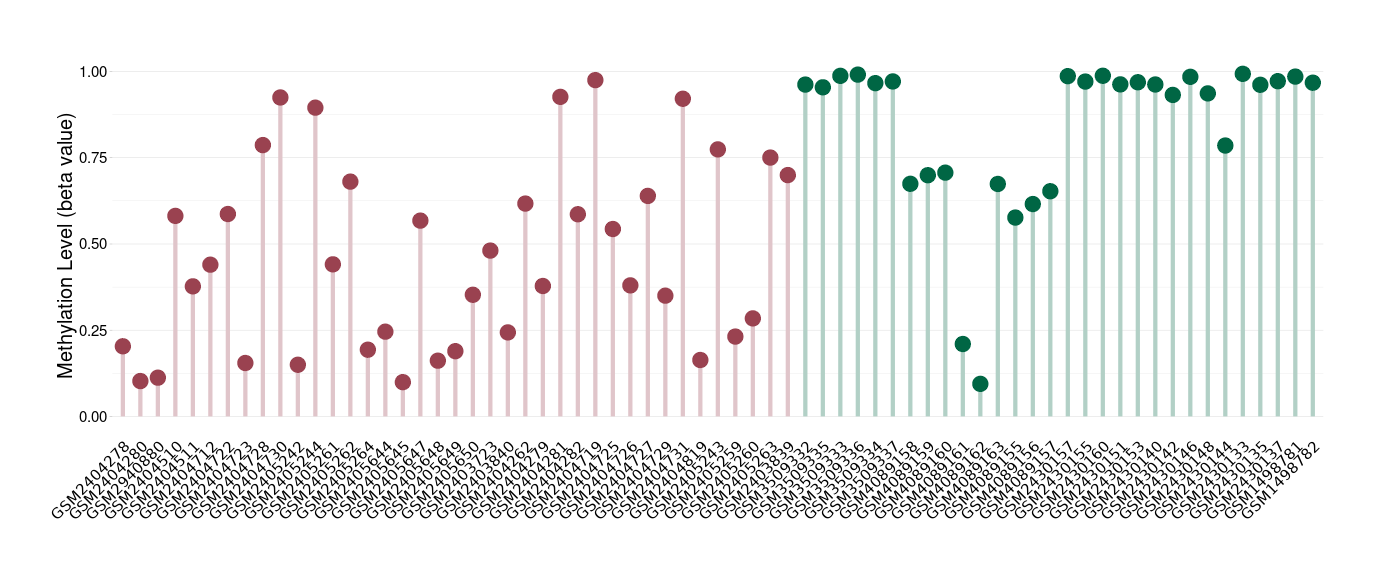 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
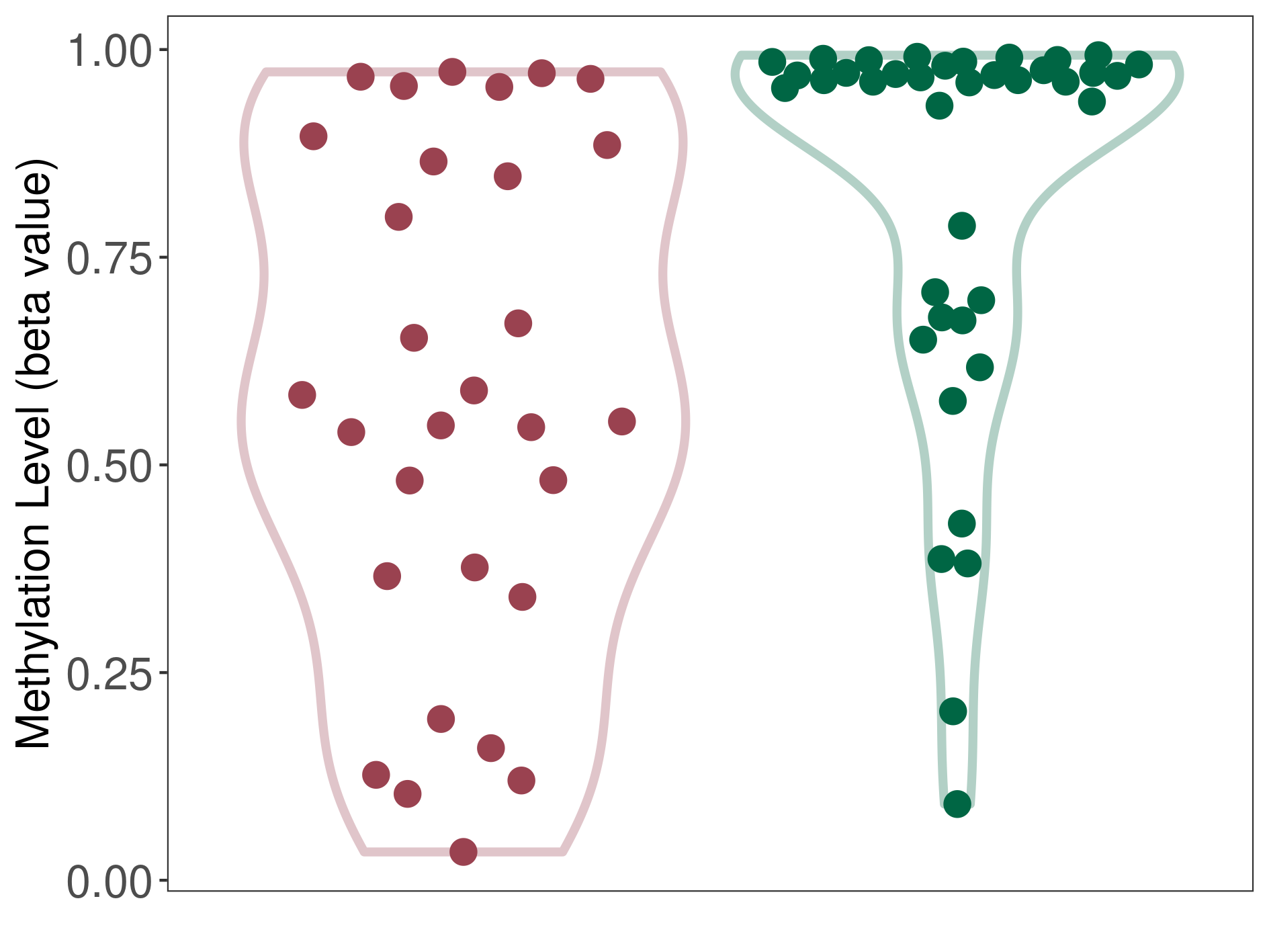
Melanoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypomethylation of KCNH7 in melanoma than that in healthy individual | ||||
Studied Phenotype |
Melanoma [ICD-11:2C30] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 0.000376238; Fold-change: -0.413113409; Z-score: -2.046396421 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
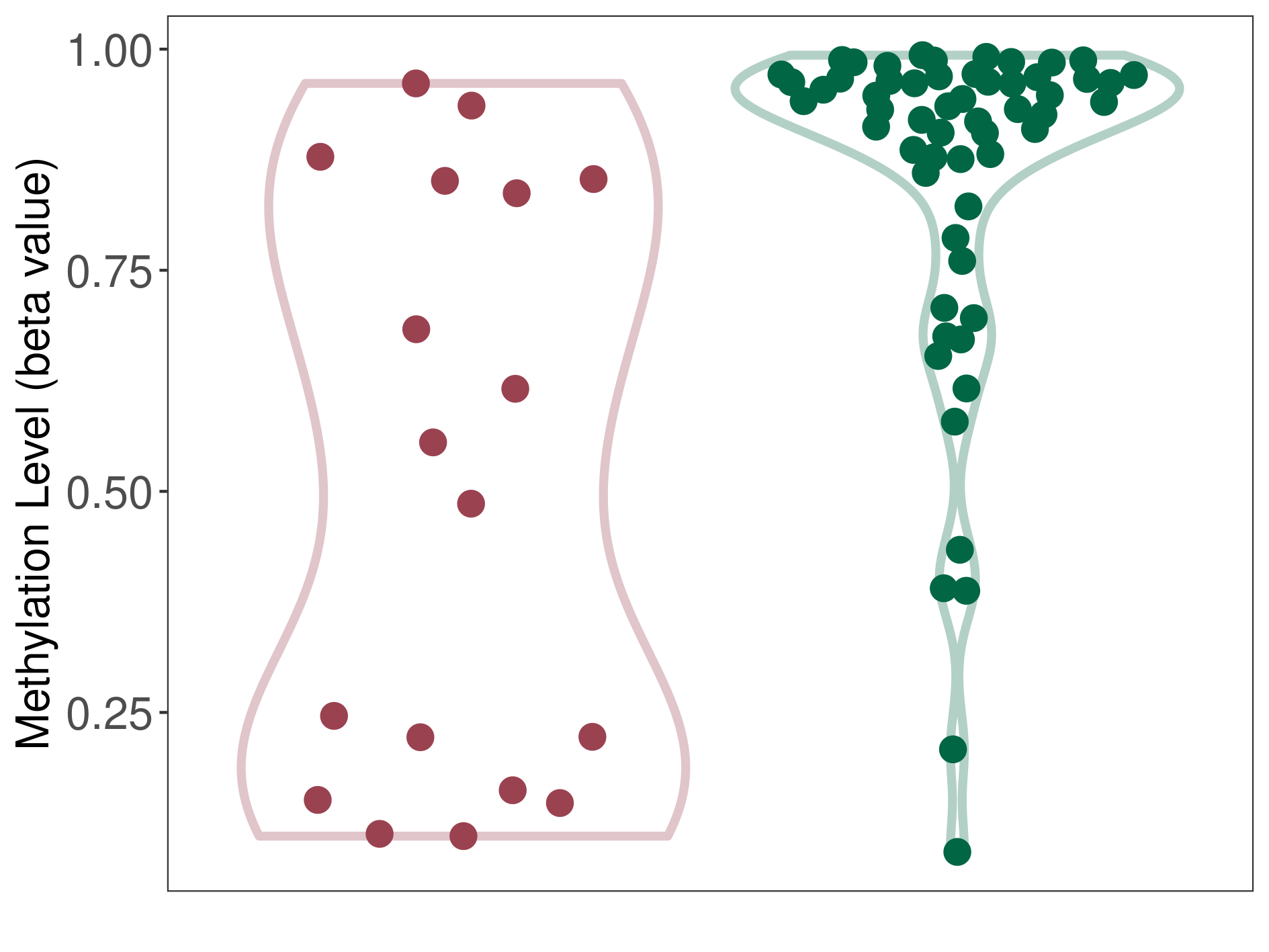
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
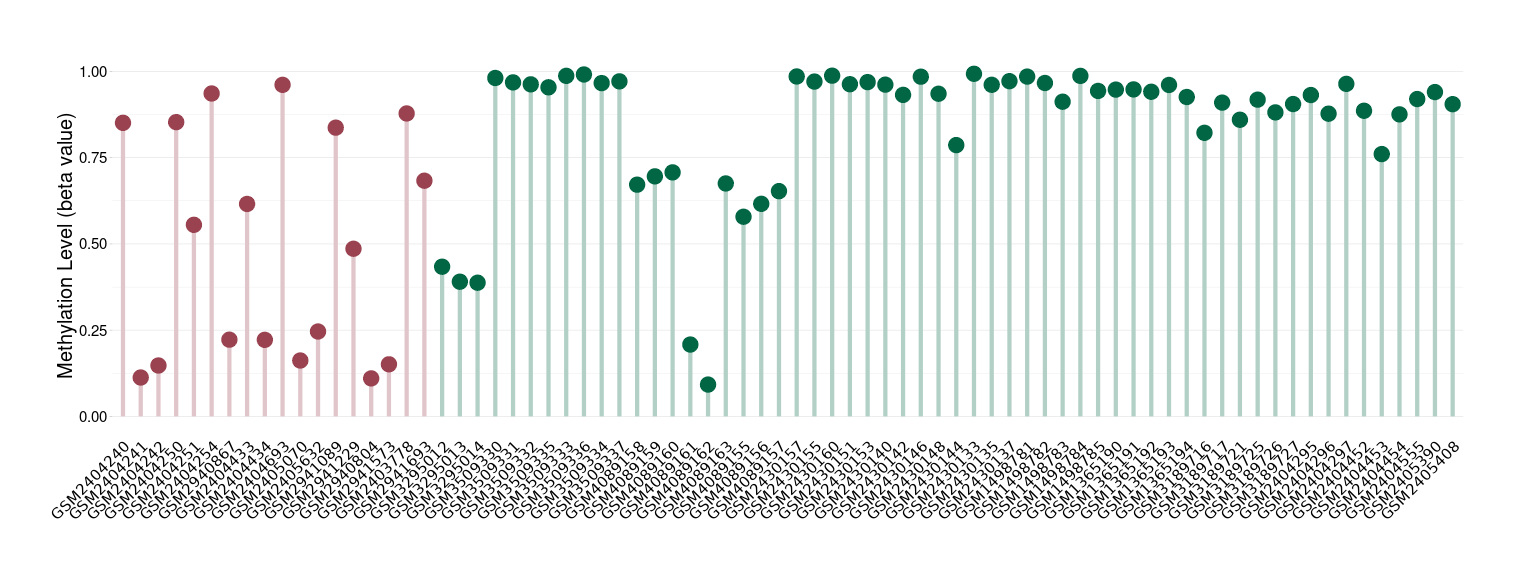
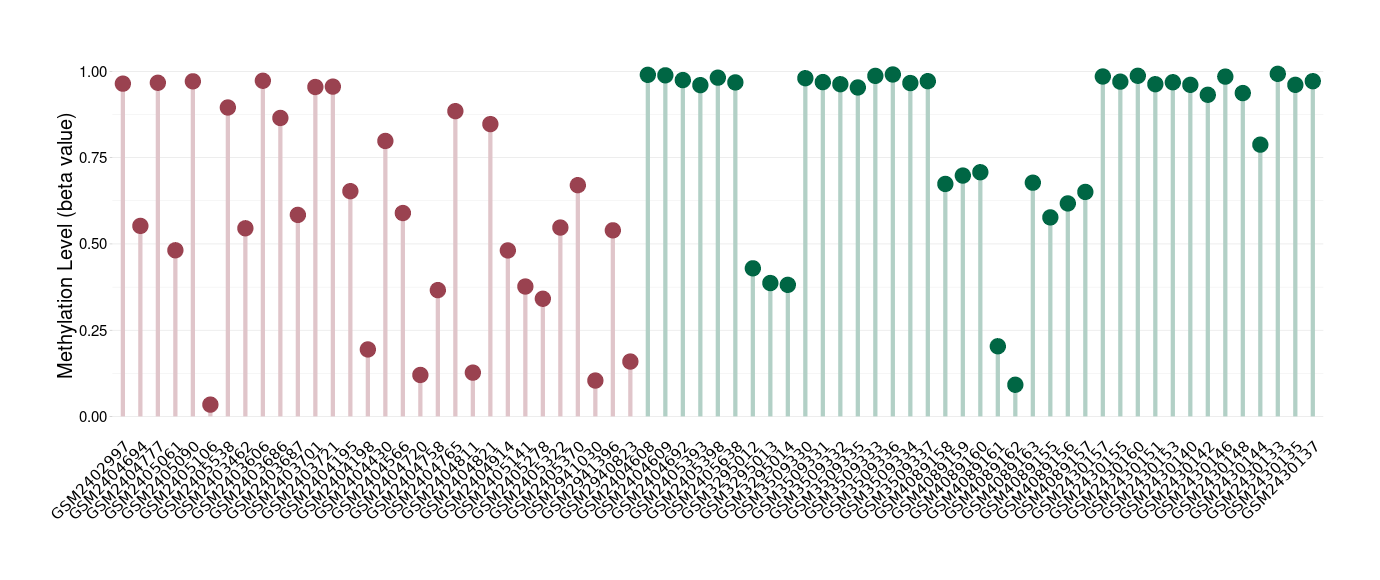
Peripheral neuroectodermal tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypomethylation of KCNH7 in peripheral neuroectodermal tumour than that in healthy individual | ||||
Studied Phenotype |
Peripheral neuroectodermal tumour [ICD-11:2B52] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 0.000909948; Fold-change: -0.394861922; Z-score: -1.623924621 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.
