Modelled Stucture
Method: Homology modeling
Teplate PDB: 3SYA_A
Identity: 99.696%
Minimized Score: -845.977
Detail: Structure Info
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0525 Transporter Info | ||||
| Gene Name | KCNJ6 | ||||
| Protein Name | Inward rectifier K(+) channel Kir3.2 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| 3D Structure |
Modelled Stucture Method: Homology modeling Teplate PDB: 3SYA_A Identity: 99.696% Minimized Score: -845.977 Detail: Structure Info |
||||
| Inter-species Structural Differences (ISD) | |||||
| Mus musculus (Mouse) | |||||
| Gene Name | Kcnj6 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure | |||||
| Click to Save PDB File in PDB Format | |||||
| Experimental Structures of This Model Oganism | |||||
| PDB ID | Scanning Method | Resolution | Expression System | Details | Ref |
| 3AT8 | X-ray | 3.3 Å | Escherichia coli | [ 1] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A | ||||
| Sequence Length | 53-380 | Mutation | No | ||
| 3AT9 | X-ray | 3.3 Å | Escherichia coli | [ 1] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A | ||||
| Sequence Length | 53-380 | Mutation | No | ||
| 3ATA | X-ray | 3.49 Å | Escherichia coli | [ 1] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A | ||||
| Sequence Length | 53-380 | Mutation | No | ||
| 3ATB | X-ray | 3.51 Å | Escherichia coli | [ 1] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A | ||||
| Sequence Length | 53-380 | Mutation | No | ||
| 3ATD | X-ray | 3.01 Å | Escherichia coli | [ 1] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A | ||||
| Sequence Length | 53-380 | Mutation | No | ||
| 3ATE | X-ray | 3.2 Å | Escherichia coli | [ 1] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A | ||||
| Sequence Length | 53-380 | Mutation | No | ||
| 3AGW | X-ray | 2.2 Å | Escherichia coli | [ 2] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A | ||||
| Sequence Length | 53-380 | Mutation | No | ||
| 3AUW | X-ray | 3.56 Å | Escherichia coli | [ 3] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A/C/B/D | ||||
| Sequence Length | 53-74; 200-380 | Mutation | No | ||
| Inter-species Structural Differences (ISD) | |||||
| Rattus norvegicus (Rat) | |||||
| Gene Name | Kcnj6 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method: Homology modeling Teplate PDB: 3SYA_A Sequence Length: 329 Identity: 99.392% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -841.161 kcal/mol | |||
| Ramachandra Favored |
Medium |
||||
| QMEANBrane Quality |
High |
||||
| Ramachandran Plot |
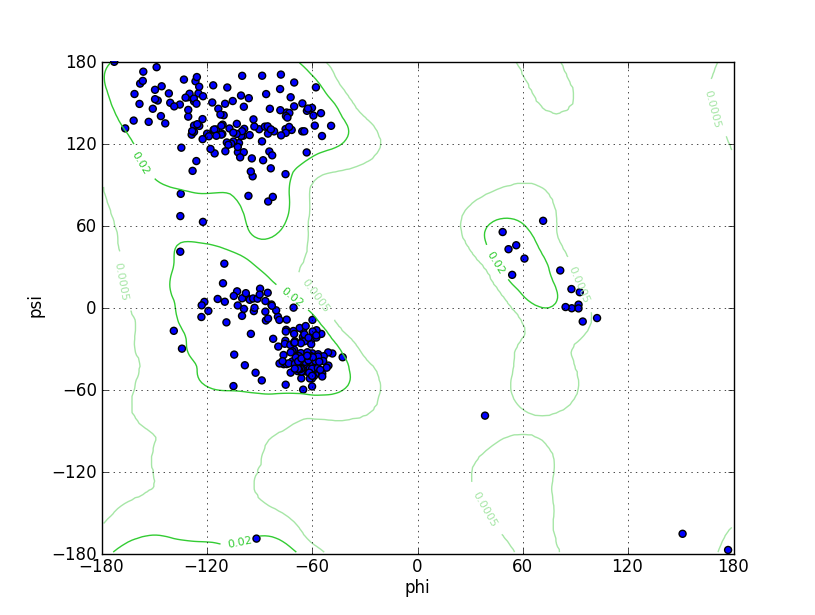
Ramz Z Score: 0.58 ± 0.45 Residues in Favored Region: 319 Ramachandran favored: 97.55% Number of Outliers: 1 Ramachandran outliers: 0.31% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
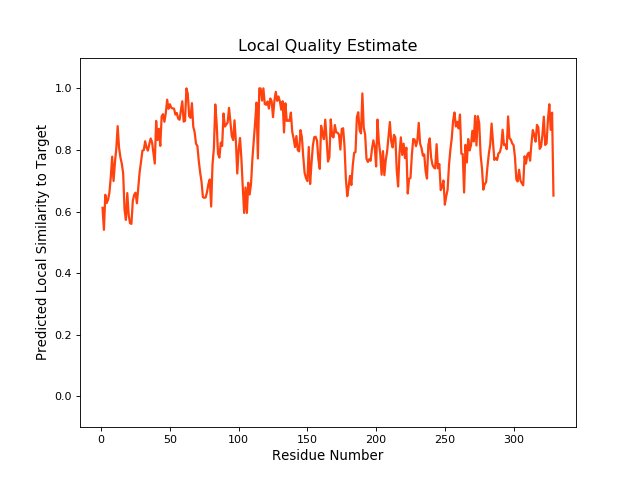
QMEANBrane Score: 0.8 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Inter-species Structural Differences (ISD) | |||||
| Pongo abelii (Sumatran orangutan) | |||||
| Gene Name | KCNJ6 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method: Homology modeling Teplate PDB: 3SYA_A Sequence Length: 329 Identity: 99.392% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -831.641 kcal/mol | |||
| Ramachandra Favored |
Medium |
||||
| QMEANBrane Quality |
High |
||||
| Ramachandran Plot |
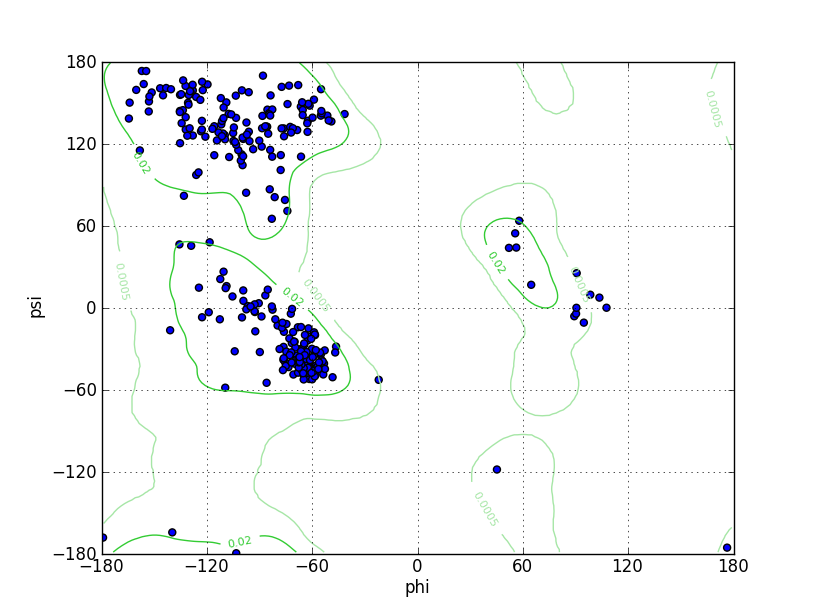
Ramz Z Score: 1.04 ± 0.45 Residues in Favored Region: 319 Ramachandran favored: 97.55% Number of Outliers: 1 Ramachandran outliers: 0.31% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
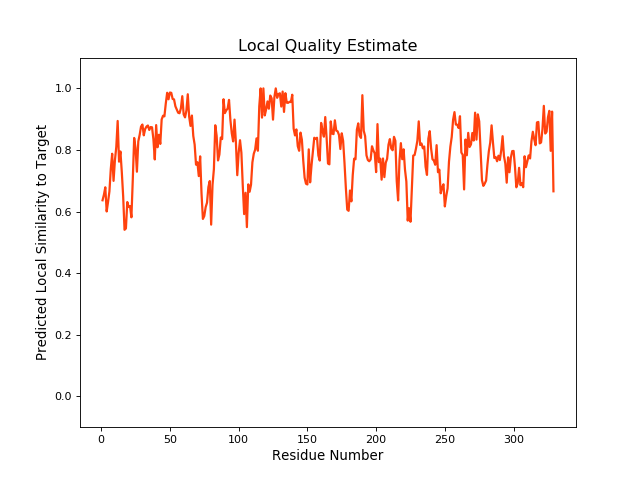
QMEANBrane Score: 0.8 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| References | |||||
| 1 | Interactions of cations with the cytoplasmic pores of inward rectifier K(+) channels in the closed state. J Biol Chem. 2011 Dec 2;286(48):41801-41811. | ||||
| 2 | A structural determinant for the control of PIP2 sensitivity in G protein-gated inward rectifier K+ channels. J Biol Chem. 2010 Dec 3;285(49):38517-23. | ||||
| 3 | Inverse agonist-like action of cadmium on G-protein-gated inward-rectifier K(+) channels. Biochem Biophys Res Commun. 2011 Apr 8;407(2):366-71. | ||||
| 4 | Crystal structure of the mammalian GIRK2 K+ channel and gating regulation by G proteins, PIP2, and sodium. Cell. 2011 Sep 30;147(1):199-208. | ||||
| 5 | X-ray structure of the mammalian GIRK2- G-protein complex. Nature. 2013 Jun 13;498(7453):190-7. | ||||
| 6 | RCSB PDB: Structural Diversity in the Cytoplasmic Region of G Protein-Gated Inward Rectifier K+ Channels. | ||||
| 7 | RCSB PDB: Coupling of G Protein Binding to Channel Gating in Mammalian Inward Rectifier K+ channels. | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.