Modelled Stucture
Method: Homology modeling
Teplate PDB: 6JPA_C
Identity: 72.951%
Minimized Score: -919.207
Detail: Structure Info
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0535 Transporter Info | ||||
| Gene Name | CACNB2 | ||||
| Protein Name | Voltage-dependent L-type calcium channel beta-2 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| 3D Structure |
Modelled Stucture Method: Homology modeling Teplate PDB: 6JPA_C Identity: 72.951% Minimized Score: -919.207 Detail: Structure Info |
||||
| Inter-species Structural Differences (ISD) | |||||
| Mus musculus (Mouse) | |||||
| Gene Name | Cacnb2 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method: Homology modeling Teplate PDB: 6JPA_C Sequence Length: 439 Identity: 77.13% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -829.583 kcal/mol | |||
| Ramachandra Favored |
Medium |
||||
| QMEANBrane Quality |
Medium |
||||
| Ramachandran Plot |
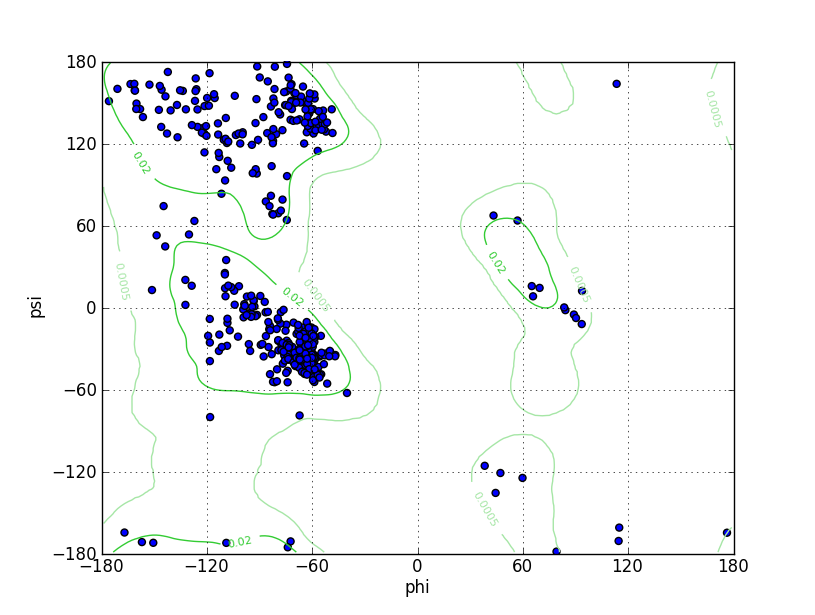
Ramz Z Score: -0.78 ± 0.4 Residues in Favored Region: 420 Ramachandran favored: 96.11% Number of Outliers: 1 Ramachandran outliers: 0.23% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
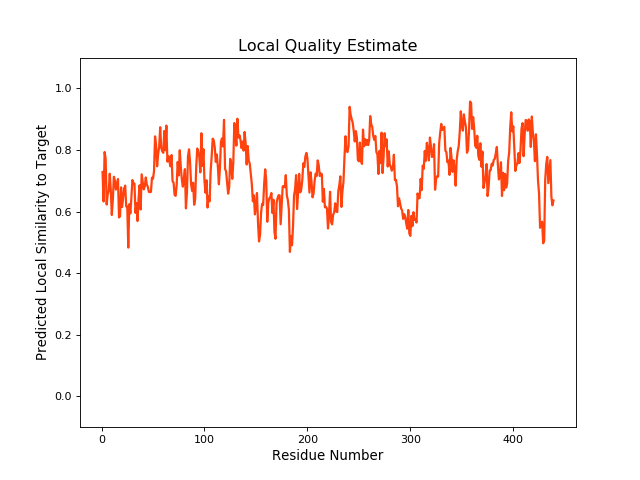
QMEANBrane Score: 0.73 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Inter-species Structural Differences (ISD) | |||||
| Rattus norvegicus (Rat) | |||||
| Gene Name | Cacnb2 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure | |||||
| Click to Save PDB File in PDB Format | |||||
| Experimental Structures of This Model Oganism | |||||
| PDB ID | Scanning Method | Resolution | Expression System | Details | Ref |
| 1T0H | X-ray | 1.97 Å | Escherichia coli | [ 1] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A/B | ||||
| Sequence Length | 68-196; 254-476 | Mutation | Yes | ||
| 5V2P | X-ray | 2 Å | Escherichia coli | [ 2] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A | ||||
| Sequence Length | 68-189; 254-476 | Mutation | Yes | ||
| 5V2Q | X-ray | 1.7 Å | Escherichia coli | [ 2] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A | ||||
| Sequence Length | 68-189; 254-476 | Mutation | Yes | ||
| 3JBR | EM | 4.2 Å | Escherichia coli | [ 3] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | B | ||||
| Sequence Length | 68-196; 254-475 | Mutation | No | ||
| Inter-species Structural Differences (ISD) | |||||
| Oryctolagus cuniculus (Rabbit) | |||||
| Gene Name | CACNB2 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method: Homology modeling Teplate PDB: 6JPA_C Sequence Length: 438 Identity: 77.303% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -799.923 kcal/mol | |||
| Ramachandra Favored |
Medium |
||||
| QMEANBrane Quality |
Medium |
||||
| Ramachandran Plot |

Ramz Z Score: 0.02 ± 0.38 Residues in Favored Region: 417 Ramachandran favored: 95.64% Number of Outliers: 1 Ramachandran outliers: 0.23% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |

QMEANBrane Score: 0.73 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Experimental Structures of This Model Oganism | |||||
| PDB ID | Scanning Method | Resolution | Expression System | Details | Ref |
| 4DEX | X-ray | 2 Å | Escherichia coli | [ 4] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A | ||||
| Sequence Length | 51-163; 229-448 | Mutation | No | ||
| 4DEY | X-ray | 1.95 Å | Escherichia coli | [ 4] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A | ||||
| Sequence Length | 51-163; 229-448 | Mutation | No | ||
| 1T3L | X-ray | 2.2 Å | Escherichia coli | [ 5] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A | ||||
| Sequence Length | 51-163; 229-448 | Mutation | Yes | ||
| 1T3S | X-ray | 2.3 Å | Escherichia coli | [ 5] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A | ||||
| Sequence Length | 51-163; 229-422 | Mutation | Yes | ||
| References | |||||
| 1 | Structure of a complex between a voltage-gated calcium channel beta-subunit and an alpha-subunit domain. Nature. 2004 Jun 10;429(6992):671-5. | ||||
| 2 | Stapled Voltage-Gated Calcium Channel (Ca(V)) -Interaction Domain (AID) Peptides Act As Selective Protein-Protein Interaction Inhibitors of Ca(V) Function. ACS Chem Neurosci. 2017 Jun 21;8(6):1313-1326. | ||||
| 3 | Structure of the voltage-gated calcium channel Cav1.1 complex. Science. 2015 Dec 18;350(6267):aad2395. | ||||
| 4 | The role of a voltage-dependent Ca2+ channel intracellular linker: a structure-function analysis. J Neurosci. 2012 May 30;32(22):7602-13. | ||||
| 5 | Structural analysis of the voltage-dependent calcium channel beta subunit functional core and its complex with the alpha 1 interaction domain. Neuron. 2004 May 13;42(3):387-99. | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.