Detail Information of Epigenetic Regulations
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0004 Transporter Info | ||||
| Gene Name | ABCG2 | ||||
| Transporter Name | Breast cancer resistance protein | ||||
| Gene ID | |||||
| UniProt ID | |||||
| Epigenetic Regulations of This DT (EGR) | |||||
|---|---|---|---|---|---|
|
Methylation |
|||||
|
Pancreatic cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Hypomethylation of ABCG2 in pancreatic cancer | [ 1 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Bisulfite sequencing | ||
|
Related Molecular Changes |
Up regulation of ABCG2 | Experiment Method | RT-qPCR | ||
|
Studied Phenotype |
Pancreatic cancer [ ICD-11: 2C10] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
ABCG2 expression correlated with pancreatic cancer tumorigenesis and drug resistance in a mechanism that is independent of promoter methylation. | ||||
|
Chronic myelogenous leukemia |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Hypermethylation of ABCG2 in chronic myelogenous leukemia | [ 2 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Methylation-sensitive PCR | ||
|
Related Molecular Changes |
Down regulation of ABCG2 | Experiment Method | Southern blotting | ||
|
Studied Phenotype |
Chronic myelogenous leukemia [ ICD-11: 2A20.0] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
c-MYCmediated transcription of ABCG2 gene is controlled by its CpG islandpromoter methylation status. | ||||
|
Small cell lung cancer |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Hypermethylation of ABCG2 in lung cancer (compare with drug-resistant counterpart cells) | [ 3 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Methylation-specific PCR | ||
|
Related Molecular Changes |
Down regulation of ABCG2 | Experiment Method | RT-qPCR | ||
|
Studied Phenotype |
Small cell lung cancer [ ICD-11: 2C25.1] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
Methylation status of ABCG2 is correlated inversely with its expression in drug-resistant lung cancer cells. | ||||
|
Epigenetic Phenomenon 2 |
Hypermethylation of ABCG2 in lung cancer (compare with drug-resistant counterpart cells) | [ 3 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Methylation-specific PCR | ||
|
Related Molecular Changes |
Down regulation of ABCG2 | Experiment Method | Western Blot | ||
|
Studied Phenotype |
Small cell lung cancer [ ICD-11: 2C25.1] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
Methylation status of ABCG2 is correlated inversely with its expression in drug-resistant lung cancer cells. | ||||
|
Multiple myeloma |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Hypermethylation of ABCG2 in multiple myeloma | [ 4 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Methylation-specific PCR | ||
|
Related Molecular Changes |
Down regulation of ABCG2 | Experiment Method | RT-qPCR | ||
|
Studied Phenotype |
Multiple myeloma [ ICD-11: 2A83] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
Demethylation of the promoter increased ABCG2 mRNA and protein expression. | ||||
|
Epigenetic Phenomenon 2 |
Hypermethylation of ABCG2 in multiple myeloma | [ 4 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Methylation-specific PCR | ||
|
Related Molecular Changes |
Down regulation of ABCG2 | Experiment Method | Western Blot | ||
|
Studied Phenotype |
Multiple myeloma [ ICD-11: 2A83] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
Demethylation of the promoter increased ABCG2 mRNA and protein expression. | ||||
|
Renal cell carcinoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Hypermethylation of ABCG2 in renal carcinoma | [ 5 ] | |||
|
Location |
Promoter (-599 to +329 bp) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Bisulfite sequencing | ||
|
Related Molecular Changes |
Down regulation of ABCG2 | Experiment Method | Semiquantitative RT-PCR | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
Methylation of the ABCG2 promoter inhibited transcriptional activity. | ||||
|
Glioblastoma multiforme |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Hypermethylation of ABCG2 in glioblastoma multiforme | [ 6 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Methylation-specific PCR | ||
|
Related Molecular Changes |
level of ABCG2 Unchanged | Experiment Method | RT-qPCR | ||
|
Studied Phenotype |
Glioblastoma multiforme [ ICD-11: 2A00.00] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Additional Notes |
No association between ABCG2 promoter methylation and ABCG2 expression or overall survival was seen. | ||||
|
Atypical teratoid rhabdoid tumor |
4 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of ABCG2 in atypical teratoid rhabdoid tumor | [ 7 ] | |||
|
Location |
5'UTR (cg02016771) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.06E+00 | Statistic Test | p-value: 1.18E-08; Z-score: -1.25E+00 | ||
|
Methylation in Case |
7.75E-01 (Median) | Methylation in Control | 8.23E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of ABCG2 in atypical teratoid rhabdoid tumor | [ 7 ] | |||
|
Location |
5'UTR (cg05919690) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.75E+00 | Statistic Test | p-value: 4.99E-08; Z-score: 1.51E+00 | ||
|
Methylation in Case |
8.62E-02 (Median) | Methylation in Control | 4.92E-02 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of ABCG2 in atypical teratoid rhabdoid tumor | [ 7 ] | |||
|
Location |
5'UTR (cg15443907) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.05E+00 | Statistic Test | p-value: 1.19E-06; Z-score: -1.09E+00 | ||
|
Methylation in Case |
8.21E-01 (Median) | Methylation in Control | 8.60E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of ABCG2 in atypical teratoid rhabdoid tumor | [ 7 ] | |||
|
Location |
Body (cg04064583) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.09E+00 | Statistic Test | p-value: 5.50E-04; Z-score: -4.51E-01 | ||
|
Methylation in Case |
4.65E-01 (Median) | Methylation in Control | 5.07E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Bladder cancer |
6 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of ABCG2 in bladder cancer | [ 8 ] | |||
|
Location |
5'UTR (cg02016771) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -2.36E+00 | Statistic Test | p-value: 2.42E-09; Z-score: -9.59E+00 | ||
|
Methylation in Case |
3.19E-01 (Median) | Methylation in Control | 7.54E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of ABCG2 in bladder cancer | [ 8 ] | |||
|
Location |
5'UTR (cg15443907) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.02E+00 | Statistic Test | p-value: 3.60E-02; Z-score: -7.05E-01 | ||
|
Methylation in Case |
8.77E-01 (Median) | Methylation in Control | 8.93E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of ABCG2 in bladder cancer | [ 8 ] | |||
|
Location |
TSS1500 (cg24352530) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -3.96E+00 | Statistic Test | p-value: 1.17E-05; Z-score: -5.36E+00 | ||
|
Methylation in Case |
8.59E-02 (Median) | Methylation in Control | 3.40E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of ABCG2 in bladder cancer | [ 8 ] | |||
|
Location |
TSS1500 (cg11000292) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -2.39E+00 | Statistic Test | p-value: 5.90E-03; Z-score: -2.04E+00 | ||
|
Methylation in Case |
3.05E-02 (Median) | Methylation in Control | 7.31E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of ABCG2 in bladder cancer | [ 8 ] | |||
|
Location |
TSS200 (cg26489994) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.81E+00 | Statistic Test | p-value: 3.96E-03; Z-score: -2.69E+00 | ||
|
Methylation in Case |
1.89E-02 (Median) | Methylation in Control | 3.42E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of ABCG2 in bladder cancer | [ 8 ] | |||
|
Location |
Body (cg04064583) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.92E+00 | Statistic Test | p-value: 1.37E-04; Z-score: -3.80E+00 | ||
|
Methylation in Case |
6.39E-02 (Median) | Methylation in Control | 1.23E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Breast cancer |
9 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of ABCG2 in breast cancer | [ 9 ] | |||
|
Location |
5'UTR (cg15443907) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.03E+00 | Statistic Test | p-value: 8.04E-04; Z-score: -9.54E-01 | ||
|
Methylation in Case |
8.80E-01 (Median) | Methylation in Control | 9.05E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of ABCG2 in breast cancer | [ 9 ] | |||
|
Location |
5'UTR (cg02016771) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.09E+00 | Statistic Test | p-value: 2.29E-03; Z-score: -8.10E-01 | ||
|
Methylation in Case |
6.82E-01 (Median) | Methylation in Control | 7.45E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of ABCG2 in breast cancer | [ 9 ] | |||
|
Location |
5'UTR (cg05919690) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.02E+00 | Statistic Test | p-value: 3.56E-02; Z-score: -3.32E-01 | ||
|
Methylation in Case |
6.67E-01 (Median) | Methylation in Control | 6.82E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of ABCG2 in breast cancer | [ 9 ] | |||
|
Location |
TSS1500 (cg11000292) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.07E+00 | Statistic Test | p-value: 1.13E-02; Z-score: 1.82E-01 | ||
|
Methylation in Case |
3.61E-02 (Median) | Methylation in Control | 3.37E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of ABCG2 in breast cancer | [ 9 ] | |||
|
Location |
TSS1500 (cg27493371) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.05E+00 | Statistic Test | p-value: 3.71E-02; Z-score: 3.45E-01 | ||
|
Methylation in Case |
1.68E-01 (Median) | Methylation in Control | 1.59E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of ABCG2 in breast cancer | [ 9 ] | |||
|
Location |
TSS200 (cg03415858) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.10E+00 | Statistic Test | p-value: 7.24E-03; Z-score: 3.28E-01 | ||
|
Methylation in Case |
5.32E-02 (Median) | Methylation in Control | 4.83E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of ABCG2 in breast cancer | [ 9 ] | |||
|
Location |
TSS200 (cg26489994) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.16E+00 | Statistic Test | p-value: 1.47E-02; Z-score: 1.05E-01 | ||
|
Methylation in Case |
1.79E-02 (Median) | Methylation in Control | 1.54E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of ABCG2 in breast cancer | [ 9 ] | |||
|
Location |
TSS200 (cg25295218) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.64E+00 | Statistic Test | p-value: 3.85E-02; Z-score: 4.58E-01 | ||
|
Methylation in Case |
2.71E-02 (Median) | Methylation in Control | 1.65E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of ABCG2 in breast cancer | [ 9 ] | |||
|
Location |
Body (cg04064583) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.61E+00 | Statistic Test | p-value: 1.25E-06; Z-score: -1.66E+00 | ||
|
Methylation in Case |
1.87E-01 (Median) | Methylation in Control | 3.01E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Hepatocellular carcinoma |
6 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of ABCG2 in hepatocellular carcinoma | [ 10 ] | |||
|
Location |
5'UTR (cg15443907) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.03E+00 | Statistic Test | p-value: 4.49E-03; Z-score: -6.16E-01 | ||
|
Methylation in Case |
8.44E-01 (Median) | Methylation in Control | 8.66E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of ABCG2 in hepatocellular carcinoma | [ 10 ] | |||
|
Location |
5'UTR (cg02016771) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.00E+00 | Statistic Test | p-value: 1.69E-02; Z-score: -5.10E-03 | ||
|
Methylation in Case |
8.32E-01 (Median) | Methylation in Control | 8.33E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of ABCG2 in hepatocellular carcinoma | [ 10 ] | |||
|
Location |
TSS1500 (cg24352530) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.38E+00 | Statistic Test | p-value: 2.61E-03; Z-score: -1.33E+00 | ||
|
Methylation in Case |
9.47E-02 (Median) | Methylation in Control | 1.30E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of ABCG2 in hepatocellular carcinoma | [ 10 ] | |||
|
Location |
Body (cg20080616) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.53E+00 | Statistic Test | p-value: 1.91E-14; Z-score: -5.48E+00 | ||
|
Methylation in Case |
5.09E-01 (Median) | Methylation in Control | 7.80E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of ABCG2 in hepatocellular carcinoma | [ 10 ] | |||
|
Location |
Body (cg05369268) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.17E+00 | Statistic Test | p-value: 5.10E-11; Z-score: -4.61E+00 | ||
|
Methylation in Case |
8.25E-01 (Median) | Methylation in Control | 9.62E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of ABCG2 in hepatocellular carcinoma | [ 10 ] | |||
|
Location |
Body (cg04064583) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.24E+00 | Statistic Test | p-value: 1.92E-06; Z-score: -8.38E-01 | ||
|
Methylation in Case |
1.21E-01 (Median) | Methylation in Control | 1.50E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Panic disorder |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of ABCG2 in panic disorder | [ 11 ] | |||
|
Location |
5'UTR (cg02016771) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.04E+00 | Statistic Test | p-value: 8.39E-03; Z-score: 2.81E-01 | ||
|
Methylation in Case |
2.33E+00 (Median) | Methylation in Control | 2.25E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder [ ICD-11: 6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Renal cell carcinoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of ABCG2 in clear cell renal cell carcinoma | [ 12 ] | |||
|
Location |
TSS1500 (cg24352530) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.31E+00 | Statistic Test | p-value: 1.06E-02; Z-score: -1.20E+00 | ||
|
Methylation in Case |
1.64E-01 (Median) | Methylation in Control | 2.15E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colorectal cancer |
5 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of ABCG2 in colorectal cancer | [ 13 ] | |||
|
Location |
TSS1500 (cg02196227) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.00E+00 | Statistic Test | p-value: 3.72E-02; Z-score: -2.59E-02 | ||
|
Methylation in Case |
1.22E-01 (Median) | Methylation in Control | 1.22E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of ABCG2 in colorectal cancer | [ 13 ] | |||
|
Location |
TSS200 (cg26489994) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.13E+00 | Statistic Test | p-value: 3.86E-02; Z-score: 3.87E-01 | ||
|
Methylation in Case |
1.66E-02 (Median) | Methylation in Control | 1.47E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of ABCG2 in colorectal cancer | [ 13 ] | |||
|
Location |
TSS200 (cg25295218) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.05E+00 | Statistic Test | p-value: 4.93E-02; Z-score: 1.31E-01 | ||
|
Methylation in Case |
1.62E-02 (Median) | Methylation in Control | 1.54E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of ABCG2 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg04064583) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.17E+00 | Statistic Test | p-value: 2.96E-02; Z-score: 6.86E-01 | ||
|
Methylation in Case |
1.82E-01 (Median) | Methylation in Control | 1.55E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Depression |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of ABCG2 in depression | [ 14 ] | |||
|
Location |
TSS1500 (cg24352530) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.06E+00 | Statistic Test | p-value: 4.59E-02; Z-score: 5.87E-01 | ||
|
Methylation in Case |
1.09E-01 (Median) | Methylation in Control | 1.03E-01 (Median) | ||
|
Studied Phenotype |
Depression [ ICD-11: 6A8Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Pancretic ductal adenocarcinoma |
9 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of ABCG2 in pancretic ductal adenocarcinoma | [ 15 ] | |||
|
Location |
TSS1500 (cg24507762) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.67E+00 | Statistic Test | p-value: 1.93E-13; Z-score: 2.55E+00 | ||
|
Methylation in Case |
3.72E-01 (Median) | Methylation in Control | 2.23E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of ABCG2 in pancretic ductal adenocarcinoma | [ 15 ] | |||
|
Location |
TSS1500 (cg06213598) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.04E+00 | Statistic Test | p-value: 1.68E-02; Z-score: -5.28E-01 | ||
|
Methylation in Case |
7.99E-01 (Median) | Methylation in Control | 8.27E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of ABCG2 in pancretic ductal adenocarcinoma | [ 15 ] | |||
|
Location |
TSS200 (cg13120771) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.53E+00 | Statistic Test | p-value: 2.42E-04; Z-score: -1.17E+00 | ||
|
Methylation in Case |
9.04E-02 (Median) | Methylation in Control | 1.38E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of ABCG2 in pancretic ductal adenocarcinoma | [ 15 ] | |||
|
Location |
TSS200 (cg24084363) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.11E+00 | Statistic Test | p-value: 1.31E-03; Z-score: 4.54E-01 | ||
|
Methylation in Case |
1.03E-01 (Median) | Methylation in Control | 9.27E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of ABCG2 in pancretic ductal adenocarcinoma | [ 15 ] | |||
|
Location |
TSS200 (cg09796800) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.01E+00 | Statistic Test | p-value: 1.53E-02; Z-score: 3.57E-01 | ||
|
Methylation in Case |
9.52E-01 (Median) | Methylation in Control | 9.42E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of ABCG2 in pancretic ductal adenocarcinoma | [ 15 ] | |||
|
Location |
Body (cg00244747) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.10E+00 | Statistic Test | p-value: 4.31E-05; Z-score: -1.12E+00 | ||
|
Methylation in Case |
7.22E-01 (Median) | Methylation in Control | 7.94E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of ABCG2 in pancretic ductal adenocarcinoma | [ 15 ] | |||
|
Location |
Body (cg25453063) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.02E+00 | Statistic Test | p-value: 2.46E-03; Z-score: 4.70E-01 | ||
|
Methylation in Case |
8.71E-01 (Median) | Methylation in Control | 8.52E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of ABCG2 in pancretic ductal adenocarcinoma | [ 15 ] | |||
|
Location |
Body (cg02414785) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.04E+00 | Statistic Test | p-value: 3.72E-03; Z-score: -6.37E-01 | ||
|
Methylation in Case |
6.19E-01 (Median) | Methylation in Control | 6.43E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of ABCG2 in pancretic ductal adenocarcinoma | [ 15 ] | |||
|
Location |
Body (cg15599483) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.16E+00 | Statistic Test | p-value: 1.57E-02; Z-score: -9.46E-01 | ||
|
Methylation in Case |
7.34E-01 (Median) | Methylation in Control | 8.51E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Papillary thyroid cancer |
5 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of ABCG2 in papillary thyroid cancer | [ 16 ] | |||
|
Location |
TSS1500 (cg24352530) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.32E+00 | Statistic Test | p-value: 4.57E-05; Z-score: -1.30E+00 | ||
|
Methylation in Case |
1.18E-01 (Median) | Methylation in Control | 1.55E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of ABCG2 in papillary thyroid cancer | [ 16 ] | |||
|
Location |
TSS1500 (cg11000292) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.20E+00 | Statistic Test | p-value: 9.42E-04; Z-score: -6.16E-01 | ||
|
Methylation in Case |
2.60E-02 (Median) | Methylation in Control | 3.12E-02 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of ABCG2 in papillary thyroid cancer | [ 16 ] | |||
|
Location |
TSS200 (cg26489994) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.15E+00 | Statistic Test | p-value: 3.36E-06; Z-score: -8.46E-01 | ||
|
Methylation in Case |
5.05E-02 (Median) | Methylation in Control | 5.82E-02 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of ABCG2 in papillary thyroid cancer | [ 16 ] | |||
|
Location |
TSS200 (cg25295218) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.06E+00 | Statistic Test | p-value: 9.48E-03; Z-score: -3.98E-01 | ||
|
Methylation in Case |
5.39E-02 (Median) | Methylation in Control | 5.71E-02 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of ABCG2 in papillary thyroid cancer | [ 16 ] | |||
|
Location |
Body (cg04064583) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.14E+00 | Statistic Test | p-value: 4.19E-05; Z-score: -1.01E+00 | ||
|
Methylation in Case |
4.99E-01 (Median) | Methylation in Control | 5.68E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Systemic lupus erythematosus |
4 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of ABCG2 in systemic lupus erythematosus | [ 17 ] | |||
|
Location |
TSS1500 (cg11000292) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.10E+00 | Statistic Test | p-value: 3.53E-03; Z-score: -2.70E-01 | ||
|
Methylation in Case |
5.39E-02 (Median) | Methylation in Control | 5.92E-02 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus [ ICD-11: 4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of ABCG2 in systemic lupus erythematosus | [ 17 ] | |||
|
Location |
TSS1500 (cg24352530) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.11E+00 | Statistic Test | p-value: 4.38E-03; Z-score: -4.23E-01 | ||
|
Methylation in Case |
2.00E-01 (Median) | Methylation in Control | 2.21E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus [ ICD-11: 4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of ABCG2 in systemic lupus erythematosus | [ 17 ] | |||
|
Location |
TSS1500 (cg02196227) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.03E+00 | Statistic Test | p-value: 5.16E-03; Z-score: -1.68E-01 | ||
|
Methylation in Case |
9.74E-02 (Median) | Methylation in Control | 1.00E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus [ ICD-11: 4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of ABCG2 in systemic lupus erythematosus | [ 17 ] | |||
|
Location |
TSS200 (cg01263075) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.02E+00 | Statistic Test | p-value: 6.00E-03; Z-score: -1.22E-01 | ||
|
Methylation in Case |
8.00E-02 (Median) | Methylation in Control | 8.20E-02 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus [ ICD-11: 4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
HIV infection |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of ABCG2 in HIV infection | [ 18 ] | |||
|
Location |
TSS200 (cg03415858) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.09E+00 | Statistic Test | p-value: 1.25E-02; Z-score: 4.98E-01 | ||
|
Methylation in Case |
5.89E-02 (Median) | Methylation in Control | 5.39E-02 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of ABCG2 in HIV infection | [ 18 ] | |||
|
Location |
Body (cg04064583) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.42E+00 | Statistic Test | p-value: 5.66E-05; Z-score: 1.86E+00 | ||
|
Methylation in Case |
2.74E-01 (Median) | Methylation in Control | 1.92E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Lung adenocarcinoma |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of ABCG2 in lung adenocarcinoma | [ 19 ] | |||
|
Location |
TSS200 (cg26489994) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.19E+00 | Statistic Test | p-value: 1.27E-02; Z-score: -1.30E+00 | ||
|
Methylation in Case |
3.39E-02 (Median) | Methylation in Control | 4.03E-02 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of ABCG2 in lung adenocarcinoma | [ 19 ] | |||
|
Location |
Body (cg04064583) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.18E+00 | Statistic Test | p-value: 3.40E-02; Z-score: 1.16E+00 | ||
|
Methylation in Case |
2.06E-01 (Median) | Methylation in Control | 1.74E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colon cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of ABCG2 in colon adenocarcinoma | [ 20 ] | |||
|
Location |
Body (cg04310063) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.16E+00 | Statistic Test | p-value: 1.63E-03; Z-score: -3.79E+00 | ||
|
Methylation in Case |
6.73E-01 (Median) | Methylation in Control | 7.84E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Nodular goiter |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Hypomethylation of ABCG2 in nodular goiter | [ 42 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Methylation-sensitive PCR | ||
|
Studied Phenotype |
Nodular goiter [ ICD-11: 5A01.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Additional Notes |
Promoter methylation of ABCG2 was significantly positively associated with a family history of thyroid diseases (P < 0.05). | ||||
|
Melanocytoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
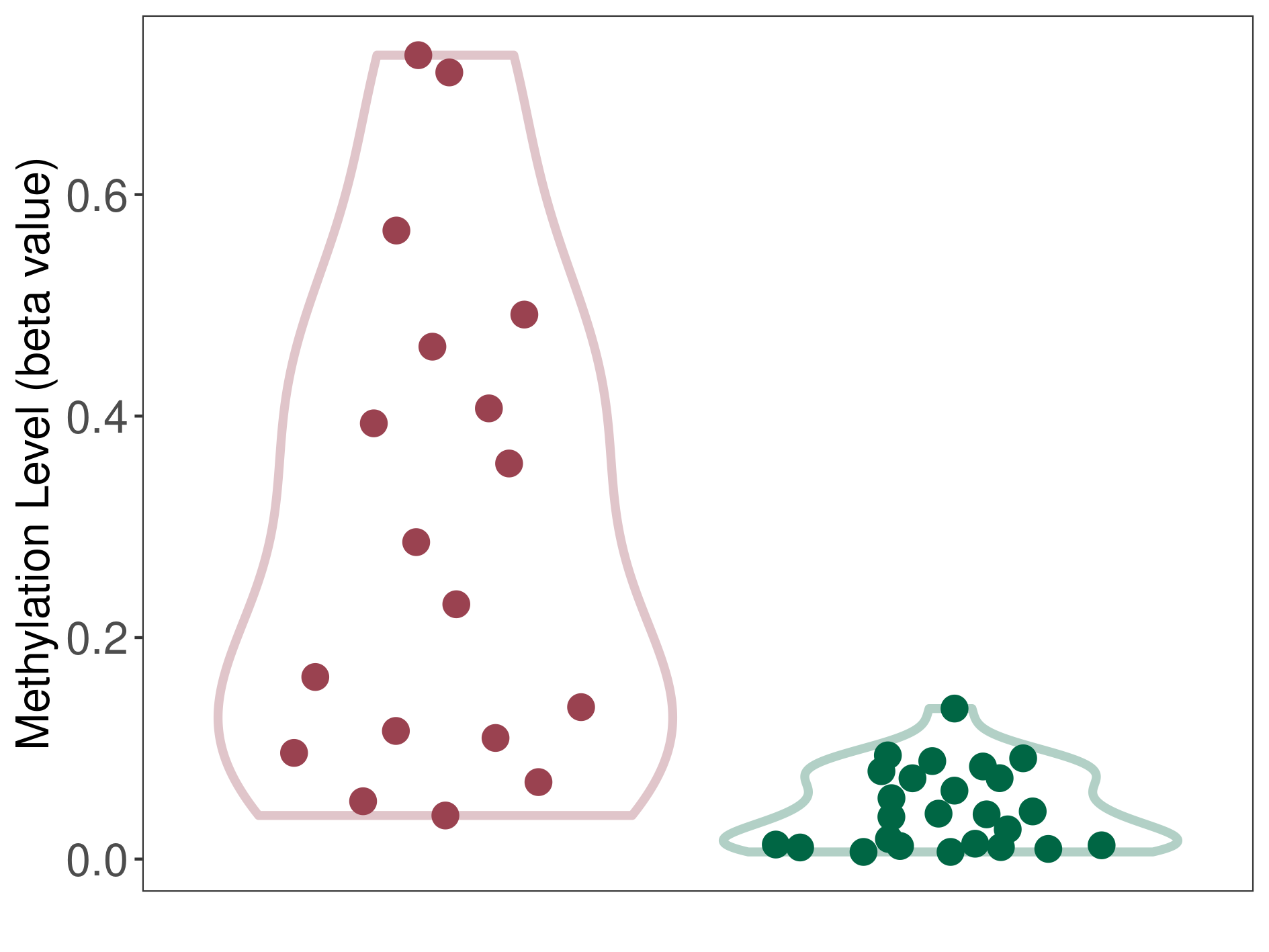
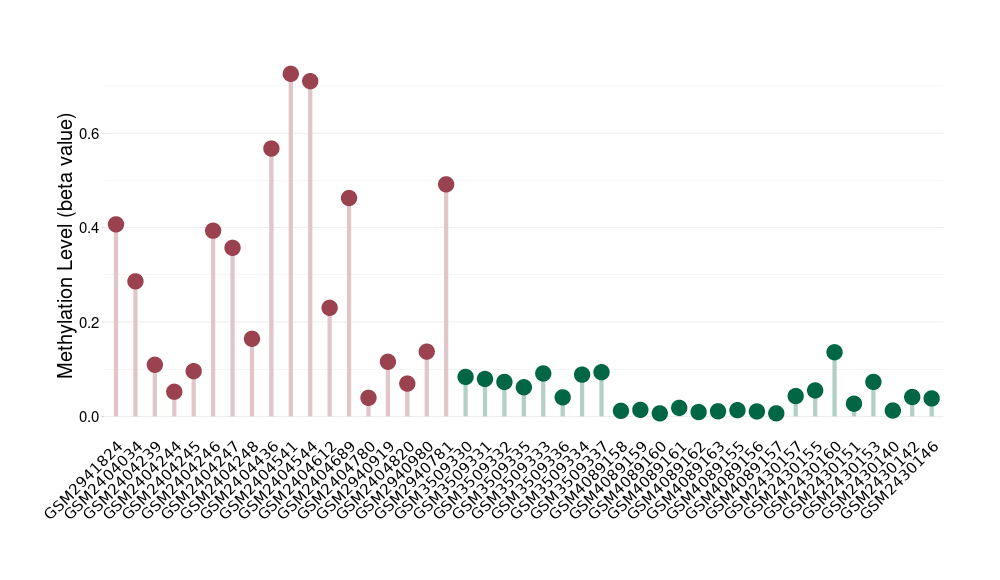
Moderate hypermethylation of ABCG2 in melanocytoma than that in healthy individual | ||||
Studied Phenotype |
Melanocytoma [ICD-11:2F36.2] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 0.000153163; Fold-change: 0.217764519; Z-score: 6.04339989 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Histone acetylation |
|||||
|
Acute myeloid leukemia |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Hypoacetylation of ABCG2 in acute myeloid leukemia (compare with valproate-treatment counterpart AML cells) | [ 21 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes |
Down regulation of ABCG2 | Experiment Method | RT-qPCR | ||
|
Studied Phenotype |
Acute myeloid leukemia [ ICD-11: 2A60] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
Chromatin immunoprecipitation revealed the?hyperacetylation?of?histone?proteins in the promoter regions of?BCRP on valproate treatment. | ||||
|
Epigenetic Phenomenon 2 |
Hypoacetylation of ABCG2 in AML (compare with valproate treatment counterpart cells) | [ 21 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes |
Down regulation of ABCG2 | Experiment Method | Western Blot | ||
|
Studied Phenotype |
Acute myeloid leukemia [ ICD-11: 2A60] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
Hyperacetylation of histone proteins in the promoter regions of BCRP on valproate treatment. | ||||
|
Epigenetic Phenomenon 3 |
Hypoacetylation of ABCG2 in AML (compare with valproate treatment counterpart cells) | [ 21 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes |
Down regulation of ABCG2 | Experiment Method | Western Blot | ||
|
Studied Phenotype |
Acute myeloid leukemia [ ICD-11: 2A60] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
Hyperacetylation of histone proteins in the promoter regions of BCRP on valproate treatment. | ||||
|
Breast cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Hypoacetylation of ABCG2 in breast cancer (compare with doxorubicin-resistant counterpart cells) | [ 22 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes |
Down regulation of ABCG2 | Experiment Method | RT-qPCR | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
High acetylation level of histone H3 of ABCG2 in doxorubicin-resistant breast cancer cells associated with higher ABCG2 expression. | ||||
|
Colon cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Hypoacetylation of ABCG2 in colon cancer (compare with romidepsin-resistant counterpart cells) | [ 23 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes |
Down regulation of ABCG2 | Experiment Method | RT-qPCR | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
High acetylation level of histone H3 of ABCG2 in romidepsin-resistant colon cancer cells associated with higher ABCG2 expression. | ||||
|
Promyeloblastic leukemia |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Hypoacetylation of ABCG2 in acute myeloid leukemia (compare with valproate-treatment counterpart AML cells) | [ 24 ] | |||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes |
Down regulation of ABCG2 | Experiment Method | RT-qPCR | ||
|
Studied Phenotype |
Promyeloblastic leukemia [ ICD-11: 2A60.0] | ||||
|
Experimental Material |
Human leukemia stem cell-like cell line (KG-1a) | ||||
|
Additional Notes |
Exposure of promyeloblastic leukemia cells for 24 h to 2 mM of the weak histone deacetylase inhibitor phenylbutyrate resulted in enhanced and marked expression of BCRP. | ||||
|
microRNA |
|||||
|
Breast cancer |
5 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Higher expression of miR-106a in breast cancer (compare with adjacent normal tissue) | [ 25 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | . | ||
|
Related Molecular Changes |
Up regulation of ABCG2 | Experiment Method | Western Blot | ||
|
miRNA Stemloop ID |
miR-106a | miRNA Mature ID | miR-106a-5p | ||
|
miRNA Sequence |
AAAAGUGCUUACAGUGCAGGUAG | ||||
|
miRNA Target Type |
Unknown | ||||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
miRNA-106a was upregulated in human breast cancer tissues. miRNA-106a promoted the proliferation, clonogenicity, migration, and invasion of breast cancer MCF-7 and MDA-MB-231 cells, and decreased their apoptosis. | ||||
|
Epigenetic Phenomenon 2 |
Higher expression of miR-181a in breast cancer (compare with mitoxantrone-resistant cells) | [ 28 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Luciferase reporter assay | ||
|
Related Molecular Changes |
Down regulation of ABCG2 | Experiment Method | RT-qPCR | ||
|
miRNA Stemloop ID |
miR-181a | miRNA Mature ID | miR-181a-5p | ||
|
miRNA Sequence |
AACAUUCAACGCUGUCGGUGAGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Human breast adenocarcinoma cell line (MCF-7) | ||||
|
Additional Notes |
miR-181a was down-regulated in mitoxantrone-resistant MCF-7/MX cells and regulated BCRP expression via binding to the 3-UTR of BCRP mRNA. | ||||
|
Epigenetic Phenomenon 3 |
Higher expression of miR-200c in breast cancer (compare with doxorubicin-resistant cells) | [ 29 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Bioinformatic prediction | ||
|
Related Molecular Changes |
Down regulation of ABCG2 | Experiment Method | RT-qPCR | ||
|
miRNA Stemloop ID |
miR-200c | miRNA Mature ID | Unclear | ||
|
miRNA Target Type |
Direct | ||||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Human breast adenocarcinoma cell line (MCF-7) | ||||
|
Additional Notes |
Down-regulation of microRNA-200c is associated with drug resistance in human breast cancer. | ||||
|
Epigenetic Phenomenon 4 |
Higher expression of miR-328 in breast cancer (compare with mitoxantrone-resistant cells) | [ 32 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Luciferase reporter assay | ||
|
Related Molecular Changes |
Down regulation of ABCG2 | Experiment Method | Western Blot | ||
|
miRNA Stemloop ID |
miR-328 | miRNA Mature ID | miR-328-3p | ||
|
miRNA Sequence |
CUGGCCCUCUCUGCCCUUCCGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Human breast adenocarcinoma cell line (MCF-7) | ||||
|
Additional Notes |
miR-328 negatively regulates ABCG2 protein expression by acting on the 3-UTR segment and that repression of ABCG2 expression via miR-328-mediated pathway is translated into significantly increased drug sensitivity in cancer cells. | ||||
|
Epigenetic Phenomenon 5 |
Higher expression of miR-487a in breast cancer (compare with mitoxantrone-resistant cells) | [ 34 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Luciferase reporter assay | ||
|
Related Molecular Changes |
Down regulation of ABCG2 | Experiment Method | Western Blot | ||
|
miRNA Stemloop ID |
miR-487a | miRNA Mature ID | miR-487a-3p | ||
|
miRNA Sequence |
AAUCAUACAGGGACAUCCAGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Human breast adenocarcinoma cell line (MCF-7) | ||||
|
Additional Notes |
miR-487a can directly regulate BCRP expression and reverse chemotherapeutic drug resistance in a subset of breast cancers. | ||||
|
Gastric cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Higher expression of miR-132 in Lgr5+ gastric cancer (compare with Lgr5- counterpart cells) | [ 26 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Luciferase reporter assay | ||
|
Related Molecular Changes |
Up regulation of ABCG2 | Experiment Method | RT-qPCR | ||
|
miRNA Stemloop ID |
miR-132 | miRNA Mature ID | miR-132-3p | ||
|
miRNA Sequence |
UAACAGUCUACAGCCAUGGUCG | ||||
|
miRNA Target Type |
Indirect(SIRT1; CREB; ABCG2) | ||||
|
Studied Phenotype |
Gastric cancer [ ICD-11: 2B72] | ||||
|
Experimental Material |
Multiple cell lines of human; Model organism in vivo (mouse) | ||||
|
Additional Notes |
The expression of miR132 was inversely correlated with SIRT1 in gastric cancer. Down-regulation of SIRT1 led to a subsequent increase of the level of acetylated CREB which in turn activated the ABCG2 signaling pathway. | ||||
|
Glioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Higher expression of miR-145 in glioma (compare with glioma stem cells) | [ 27 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Luciferase reporter assay | ||
|
Related Molecular Changes |
Down regulation of ABCG2 | Experiment Method | RT-qPCR | ||
|
miRNA Stemloop ID |
miR-145 | miRNA Mature ID | miR-145-5p | ||
|
miRNA Sequence |
GUCCAGUUUUCCCAGGAAUCCCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Studied Phenotype |
Glioma [ ICD-11: 2A00.0] | ||||
|
Experimental Material |
Chinese patient tissue samples | ||||
|
Additional Notes |
miR-145 downregulated ABCG2 expression, and miR-145-ABCG2MMP-2/9 pathway regulated the migration and invasion in GSCs. | ||||
|
Chronic myeloid leukemia |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Higher expression of miR-212 in chronic myeloid leukemia (compare with imatinib-resistant cells) | [ 30 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Luciferase reporter assay | ||
|
Related Molecular Changes |
Down regulation of ABCG2 | Experiment Method | RT-qPCR | ||
|
miRNA Stemloop ID |
miR-212 | miRNA Mature ID | Unclear | ||
|
miRNA Target Type |
Direct | ||||
|
Studied Phenotype |
Chronic myeloid leukemia [ ICD-11: 2B33.1] | ||||
|
Experimental Material |
Human immortalized myelogenous leukemia cell line(K-562) | ||||
|
Additional Notes |
Short-term and long-term imatinib treatment downregulates miR-212 expression, leading to the abolition of suppression of ABCG2 gene expression. | ||||
|
Tongue squamous cell carcinoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Higher expression of miR-212 in squamous cell carcinoma (compare with cisplatin-resistant cells) | [ 31 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Dual-luciferase reporter assay | ||
|
Related Molecular Changes |
Down regulation of ABCG2 | Experiment Method | Western Blot | ||
|
miRNA Stemloop ID |
miR-222 | miRNA Mature ID | miR-222-3p | ||
|
miRNA Sequence |
AGCUACAUCUGGCUACUGGGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Studied Phenotype |
Tongue squamous cell carcinoma [ ICD-11: 2B61.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Additional Notes |
miR-222 overexpression may increase the sensitivity of tongue squamous cell carcinoma to cisplatin by targeting ABCG2. | ||||
|
Colorectal cancer |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Higher expression of miR-328 in colorectal cancer (compare with adjacent normal tissue) | [ 33 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Luciferase reporter assay | ||
|
Related Molecular Changes |
Down regulation of ABCG2 | Experiment Method | RT-qPCR | ||
|
miRNA Stemloop ID |
miR-328 | miRNA Mature ID | miR-328-3p | ||
|
miRNA Sequence |
CUGGCCCUCUCUGCCCUUCCGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
miR-328 downregulates ABCG2 in colorectal cancer. | ||||
|
Epigenetic Phenomenon 2 |
Lower expression of miR-519c in colorectal cancer (compare with adjacent normal tissue) | [ 37 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Ribonucleoprotein immunoprecipitation | ||
|
Related Molecular Changes |
Down regulation of ABCG2 | Experiment Method | RT-qPCR | ||
|
miRNA Stemloop ID |
miR-519c | miRNA Mature ID | miR-519c-3p | ||
|
miRNA Sequence |
AAAGUGCAUCUUUUUAGAGGAU | ||||
|
miRNA Target Type |
Indirect(HuR; ABCG2) | ||||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
miR-519c downregulates ABCG2 via miR-519c/HuR/ ABCG2 pathway. | ||||
|
Prostate cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Lower expression of miR-133b in prostate cancer (compare with adjacent normal tissue) | [ 35 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Luciferase reporter assay | ||
|
Related Molecular Changes |
Down regulation of ABCG2 | Experiment Method | RT-qPCR | ||
|
miRNA Stemloop ID |
miR-133b | miRNA Mature ID | Unclear | ||
|
miRNA Target Type |
Indirect(HuR; ABCG2) | ||||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
miR-133b downregulates ABCG2 via miR-133b/HuR/ ABCG2 pathway. | ||||
|
Non-small cell lung cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Lower expression of miR-495 in non-small cell lung cancer (compare with adjacent normal tissue) | [ 36 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Luciferase reporter assay | ||
|
Related Molecular Changes |
Down regulation of ABCG2 | Experiment Method | Western Blot | ||
|
miRNA Stemloop ID |
miR-495 | miRNA Mature ID | miR-495-5p | ||
|
miRNA Sequence |
GAAGUUGCCCAUGUUAUUUUCG | ||||
|
miRNA Target Type |
Indirect(UBE2C; ABCG2) | ||||
|
Studied Phenotype |
Non-small cell lung cancer [ ICD-11: 2C25.4] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
The miR495-UBE2C-ABCG2/ERCC1 axis reverses DDP resistance via downregulation of anti-drug genes and reducing EMT in DDP-resistant NSCLC cells. | ||||
|
Colon cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
miR-142-3p downregulates ABCG2 expression in colon cancer | [ 38 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Luciferase reporter assay | ||
|
Related Molecular Changes |
Down regulation of ABCG2 | Experiment Method | RT-qPCR | ||
|
miRNA Stemloop ID |
miR-142 | miRNA Mature ID | miR-142-3p | ||
|
miRNA Sequence |
UGUAGUGUUUCCUACUUUAUGGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
OCT4 promoter is hypomethylated in colon cancer tissues, resulting in persistent low levels of miR-142-3p. The repression of miR-142-3p results in loss of inhibition of CD133, Lgr5, and ABCG2 in the cancer cells. | ||||
|
Retinoblastoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
miR-3163 downregulates ABCG2 expression | [ 39 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Luciferase reporter assay | ||
|
Related Molecular Changes |
Down regulation of ABCG2 | Experiment Method | Western Blot | ||
|
miRNA Stemloop ID |
miR-3163 | miRNA Mature ID | miR-3163 | ||
|
miRNA Sequence |
UAUAAAAUGAGGGCAGUAAGAC | ||||
|
miRNA Target Type |
Direct | ||||
|
Studied Phenotype |
Retinoblastoma [ ICD-11: 2D02.2] | ||||
|
Experimental Material |
Human retinoblastoma cell line (WERI-Rb1) | ||||
|
Additional Notes |
miR-3163 has a significant impact on ABCG2 expression and can influence proliferation, apoptosis, and drug resistance in RCSCs. | ||||
|
Hepatocellular carcinoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
miR-491 regulates ABCG2 expression in hepatocellular carcinoma | [ 40 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Luciferase reporter assay | ||
|
Related Molecular Changes |
Down regulation of ABCG2 | Experiment Method | Western Blot | ||
|
miRNA Stemloop ID |
miR-491 | miRNA Mature ID | miR-491-5p | ||
|
miRNA Sequence |
AGUGGGGAACCCUUCCAUGAGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Hepatocellular carcinoma cell line (MHCC-97L) | ||||
|
Additional Notes |
Aspirin enhanced the sensitivity of side population cells to doxorubicin via regulating the miR-491/ABCG2 signaling pathway. | ||||
|
Pancreatic cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
miR-520h regulates ABCG2 expression | [ 41 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Bioinformatic prediction | ||
|
Related Molecular Changes |
Down regulation of ABCG2 | Experiment Method | RT-qPCR | ||
|
miRNA Stemloop ID |
miR-520h | miRNA Mature ID | miR-520h | ||
|
miRNA Sequence |
ACAAAGUGCUUCCCUUUAGAGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Studied Phenotype |
Pancreatic cancer [ ICD-11: 2C10] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
miR-520h downregulates ABCG2 in pancreatic cancer cells to inhibit migration, invasion, and side populations. | ||||
|
Unclear Phenotype |
25 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
miR-10a directly targets ABCG2 | [ 43 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | CLASH | ||
|
miRNA Stemloop ID |
miR-10a | miRNA Mature ID | miR-10a-5p | ||
|
miRNA Sequence |
UACCCUGUAGAUCCGAAUUUGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon 2 |
miR-142 directly targets ABCG2 | [ 38 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Luciferase reporter assay//Western blot | ||
|
miRNA Stemloop ID |
miR-142 | miRNA Mature ID | miR-142-3p | ||
|
miRNA Sequence |
UGUAGUGUUUCCUACUUUAUGGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 3 |
miR-16-2 directly targets ABCG2 | [ 44 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-16-2 | miRNA Mature ID | miR-16-2-3p | ||
|
miRNA Sequence |
CCAAUAUUACUGUGCUGCUUUA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 4 |
miR-181a directly targets ABCG2 | [ 28 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Luciferase reporter assay//Microarray//qRT-PCR//Western blot | ||
|
miRNA Stemloop ID |
miR-181a | miRNA Mature ID | miR-181a-5p | ||
|
miRNA Sequence |
AACAUUCAACGCUGUCGGUGAGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human breast adenocarcinoma cell line (MCF-7) | ||||
|
Epigenetic Phenomenon 5 |
miR-192 directly targets ABCG2 | [ 45 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Microarray | ||
|
miRNA Stemloop ID |
miR-192 | miRNA Mature ID | miR-192-5p | ||
|
miRNA Sequence |
CUGACCUAUGAAUUGACAGCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
miR-195 directly targets ABCG2 | [ 44 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-195 | miRNA Mature ID | miR-195-3p | ||
|
miRNA Sequence |
CCAAUAUUGGCUGUGCUGCUCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 7 |
miR-222 directly targets ABCG2 | [ 31 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Luciferase reporter assay | ||
|
miRNA Stemloop ID |
miR-222 | miRNA Mature ID | miR-222-3p | ||
|
miRNA Sequence |
AGCUACAUCUGGCUACUGGGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
miR-26b directly targets ABCG2 | [ 46 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Microarray | ||
|
miRNA Stemloop ID |
miR-26b | miRNA Mature ID | miR-26b-5p | ||
|
miRNA Sequence |
UUCAAGUAAUUCAGGAUAGGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human cervical cancer cell line (Hela) | ||||
|
Epigenetic Phenomenon 9 |
miR-3167 directly targets ABCG2 | [ 44 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3167 | miRNA Mature ID | miR-3167 | ||
|
miRNA Sequence |
AGGAUUUCAGAAAUACUGGUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 10 |
miR-328 directly targets ABCG2 | [ 33 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Luciferase reporter assay//qRT-PCR//Western blot | ||
|
miRNA Stemloop ID |
miR-328 | miRNA Mature ID | miR-328-3p | ||
|
miRNA Sequence |
CUGGCCCUCUCUGCCCUUCCGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 11 |
miR-340 directly targets ABCG2 | [ 47 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-340 | miRNA Mature ID | miR-340-5p | ||
|
miRNA Sequence |
UUAUAAAGCAAUGAGACUGAUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon 12 |
miR-369 directly targets ABCG2 | [ 44 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-369 | miRNA Mature ID | miR-369-3p | ||
|
miRNA Sequence |
AAUAAUACAUGGUUGAUCUUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 13 |
miR-374a directly targets ABCG2 | [ 44 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-374a | miRNA Mature ID | miR-374a-5p | ||
|
miRNA Sequence |
UUAUAAUACAACCUGAUAAGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 14 |
miR-374b directly targets ABCG2 | [ 44 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-374b | miRNA Mature ID | miR-374b-5p | ||
|
miRNA Sequence |
AUAUAAUACAACCUGCUAAGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 15 |
miR-4539 directly targets ABCG2 | [ 47 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4539 | miRNA Mature ID | miR-4539 | ||
|
miRNA Sequence |
GCUGAACUGGGCUGAGCUGGGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon 16 |
miR-4635 directly targets ABCG2 | [ 44 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4635 | miRNA Mature ID | miR-4635 | ||
|
miRNA Sequence |
UCUUGAAGUCAGAACCCGCAA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 17 |
miR-4724 directly targets ABCG2 | [ 47 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4724 | miRNA Mature ID | miR-4724-5p | ||
|
miRNA Sequence |
AACUGAACCAGGAGUGAGCUUCG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon 18 |
miR-487a directly targets ABCG2 | [ 34 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Luciferase reporter assay//qRT-PCR//Western blot | ||
|
miRNA Stemloop ID |
miR-487a | miRNA Mature ID | miR-487a-3p | ||
|
miRNA Sequence |
AAUCAUACAGGGACAUCCAGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human breast adenocarcinoma cell line (MCF-7) | ||||
|
Epigenetic Phenomenon 19 |
miR-497 directly targets ABCG2 | [ 47 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-497 | miRNA Mature ID | miR-497-3p | ||
|
miRNA Sequence |
CAAACCACACUGUGGUGUUAGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon 20 |
miR-520h directly targets ABCG2 | [ 41 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | qRT-PCR//Western blot | ||
|
miRNA Stemloop ID |
miR-520h | miRNA Mature ID | miR-520h | ||
|
miRNA Sequence |
ACAAAGUGCUUCCCUUUAGAGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 21 |
miR-5692b directly targets ABCG2 | [ 44 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-5692b | miRNA Mature ID | miR-5692b | ||
|
miRNA Sequence |
AAUAAUAUCACAGUAGGUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 22 |
miR-5692c directly targets ABCG2 | [ 44 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-5692c | miRNA Mature ID | miR-5692c | ||
|
miRNA Sequence |
AAUAAUAUCACAGUAGGUGUAC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 23 |
miR-7158 directly targets ABCG2 | [ 47 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-7158 | miRNA Mature ID | miR-7158-3p | ||
|
miRNA Sequence |
CUGAACUAGAGAUUGGGCCCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon 24 |
miR-876 directly targets ABCG2 | [ 44 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-876 | miRNA Mature ID | miR-876-5p | ||
|
miRNA Sequence |
UGGAUUUCUUUGUGAAUCACCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 25 |
miR-889 directly targets ABCG2 | [ 44 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-889 | miRNA Mature ID | miR-889-3p | ||
|
miRNA Sequence |
UUAAUAUCGGACAACCAUUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.
