| Description of the Variability Data in VARIDT |
|---|
|
Drug transporters (DTs) are acknowledged as important determinants governing drug absorption, excretion, and, in many cases, extent of drug entry into target organs (Annu Rev Pharmacol Toxicol. 2012, 52: 249-73). The variability of DTs has thus attracted considerable attentions and broad interests (Nat Rev Drug Discov. 2015, 14: 29-44; Clin Pharmacol Ther. 2019, doi: 10.1002/cpt.1373; Ann Oncol. 2013, 24: 1513-25). There are three aspects of variability: (1) epigenetic regulations and genetic polymorphisms of DTs playing key role in drug resistance (Sci Transl Med. 2016, 8: 348ra97) and clinical treatment optimization (Adv Drug Deliv Rev. 2017, 116: 21-36); (2) species-, tissue-, and disease-specific abundances of DT proteins essential for bridging preclinical study with clinical trial (Mol Pharm. 2015, 12: 4259-69), balancing efficacy and safety (Sci Transl Med. 2016, 8: 352ra109), and predicting disease-drug interaction (Clin Pharmacol Ther. 2018, 104: 900-15), respectively; (3) exogenous factors modulating activity of DTs and leading to the altered disposition of natural substrates and the transported drugs (Adv Drug Deliv Rev. 2015, 86: 17-26; Clin Pharmacol Ther. 2016, 100: 259-67). These variability data are vital for the clinical studies, and the accumulation of such data can lay the foundation for big data-driven precision medicine. Moreover, due to the extreme complexity of drug dispositions in living organisms, the interplays among different aspects of DT variability have recently emerged to be a new promising research direction of high-impact studies (Drug Resist Updat. 2017, 32: 23-46; Cancer Lett. 2016, 370: 153-64). Particularly, one of the common mechanisms of the cancer multidrug resistance (MDR) is the overexpression of the particular DTs in cancer cells (Cancer Lett. 2016, 370: 153-64), and the developments of exogenous chemical to inhibit the efflux of these DTs are successfully proposed to reverse caner MDR (Drug Resist Updat. 2017, 32: 23-46; Cancer Lett. 2016, 370: 153-64). These and other potential studies require the collectively analyses of multiple data among different aspects of DT variability. However, no database has been available so far to provide the comprehensive information of all aspects of DT variability. Therefore, the Variability of Drug Transporter Database (VARIDT) was introduced. First, a comprehensive literature review on all drugs approved by U.S. FDA and >1,000 drugs in clinical trial or preclinical research was conducted. Different from the small amount of DTs (~20) well characterized in previous work (Nat Rev Drug Discov. 2010, 9: 215-36), 177 DTs were confirmed for the first time in VARIDT to transport approved drug(s) and 146 DTs were to transport the drug(s) in clinical/preclinical test. Second, for these hundreds of newly confirmed DTs, the VARIDT not only comprehensively provided all three aspect of their variability data but also allowed the mutual connection (interplay) between any two aspects. All in all, the VARIDT is UNIQUE in (1) covering the largest number of DTs confirmed by their corresponding drugs among available knowledge bases, (2) providing for the first time the comprehensive sets of DT variability data, and (3) allowing the interplay analysis among different aspects of DT variability. All diseases in VARIDT were standardized using the latest revision of International Classification of Diseases (ICD-11) to serve comprehensive health management, and the vast majority (~80%) of the confirmed DTs in VARIDT had the data from multiple aspects of variability, which allowed interplays among these aspects. All data are FULLY DOWNLOADABLE . |
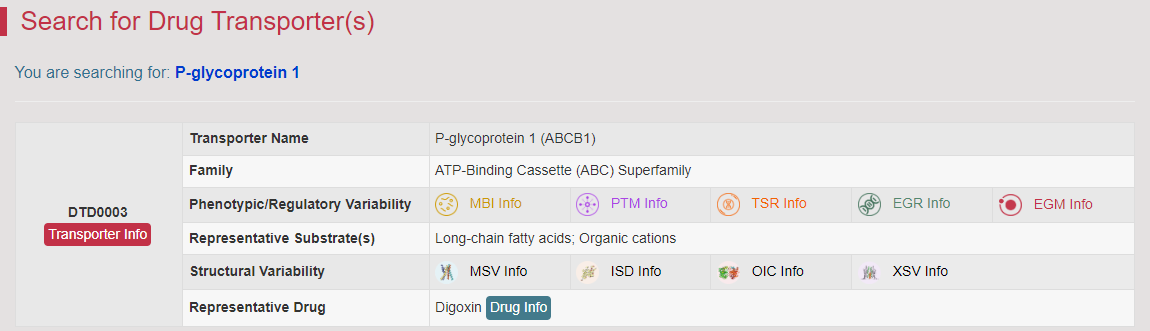
How to Search for Drug Transporter and Variability Data in VARIDT?
Drug Transporter (DT)
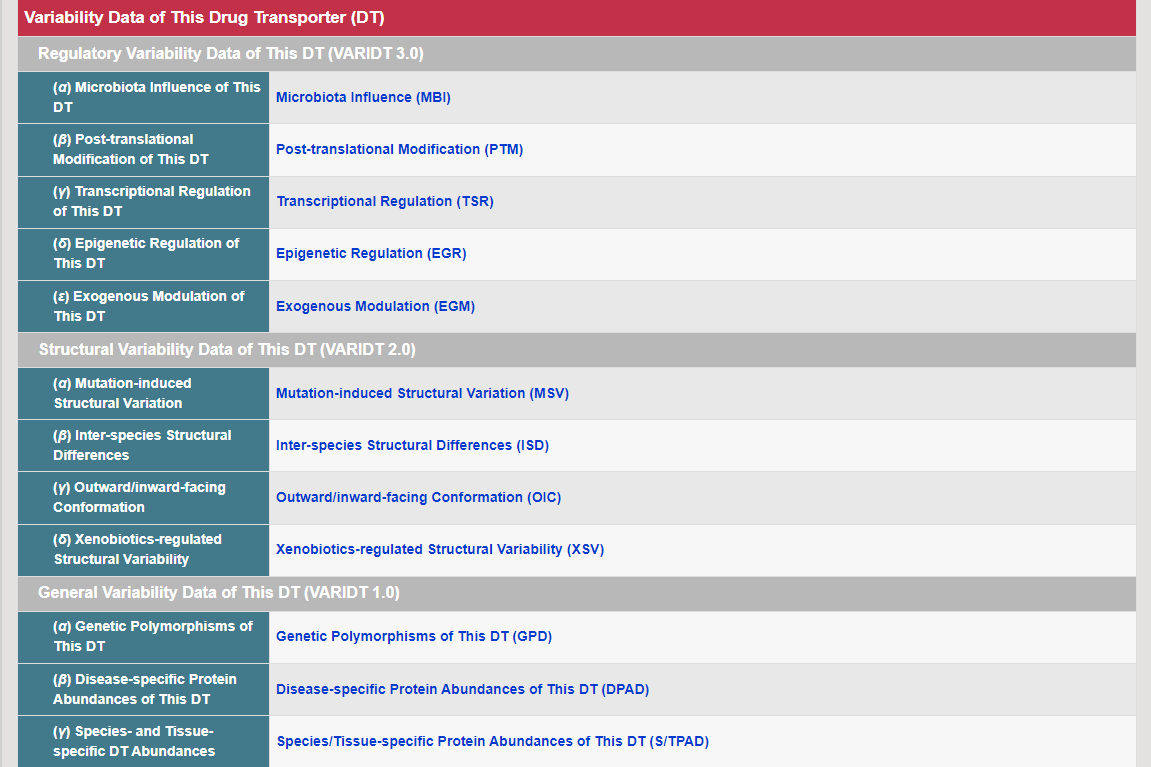
Regulatory Variability (2024 Update)
| Regulatory Variability (2024 Update) |
|---|
|
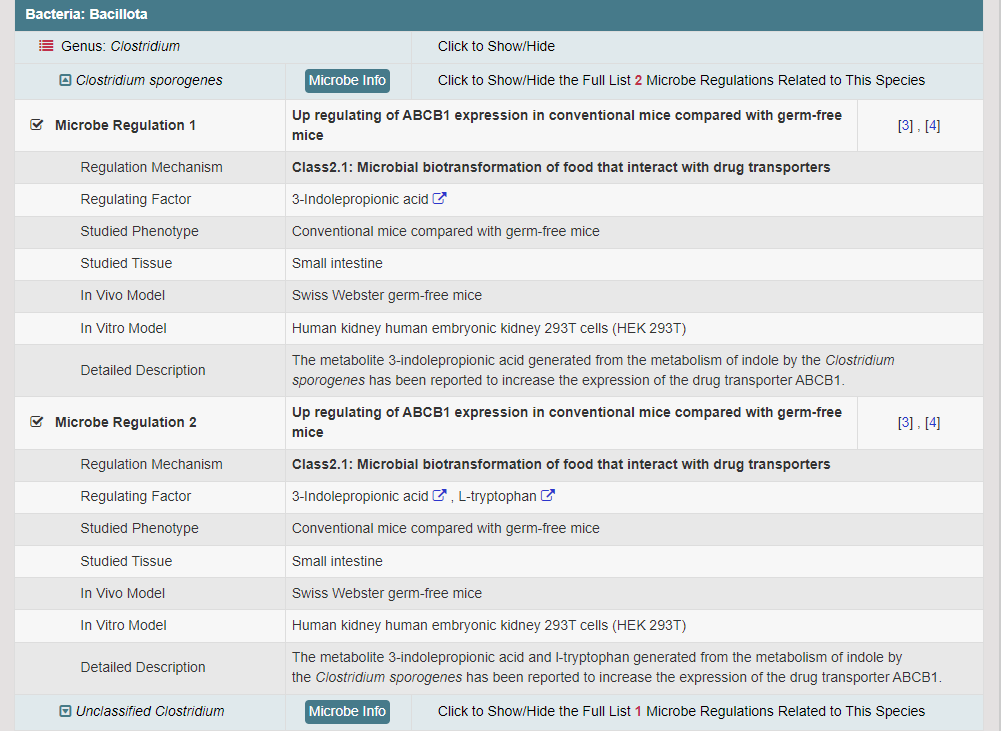
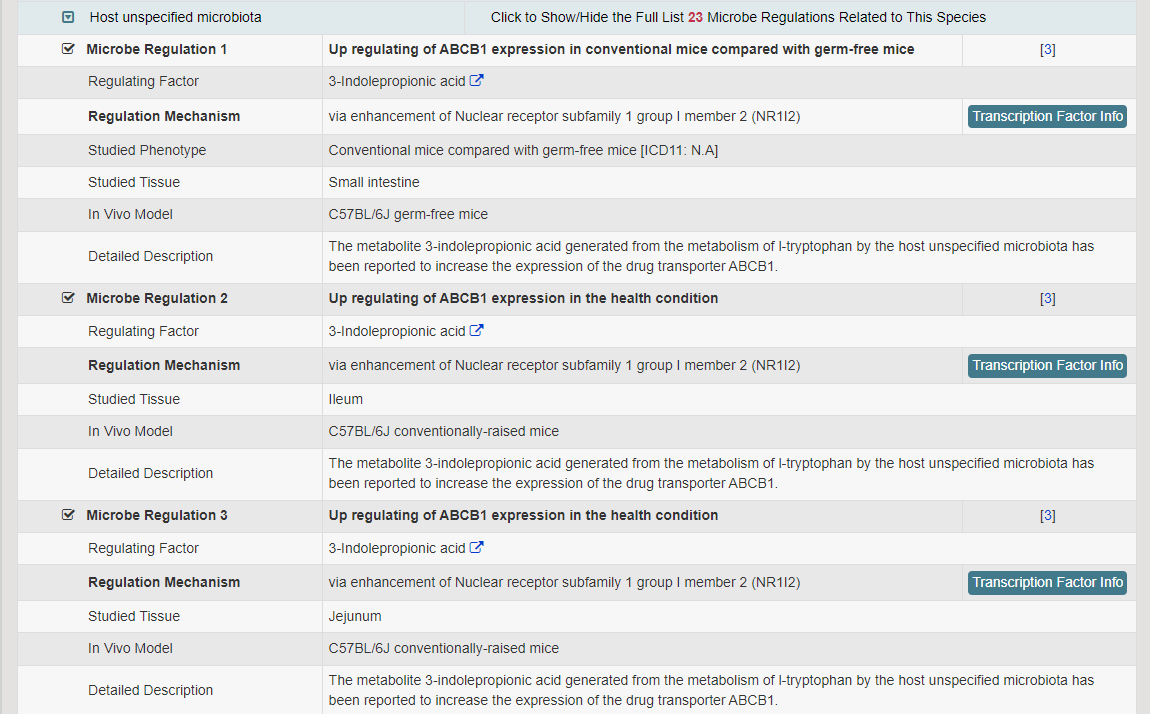
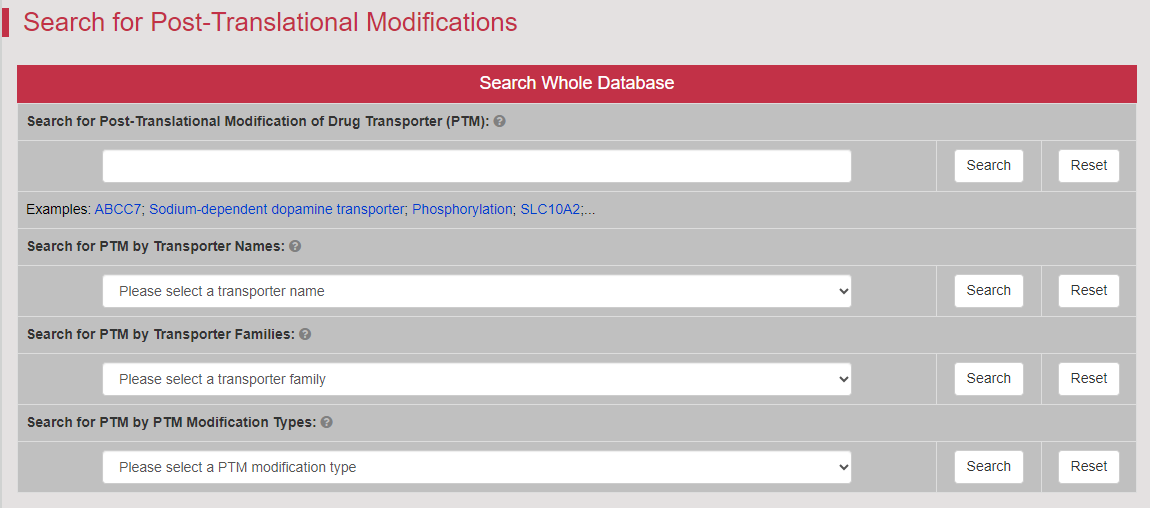
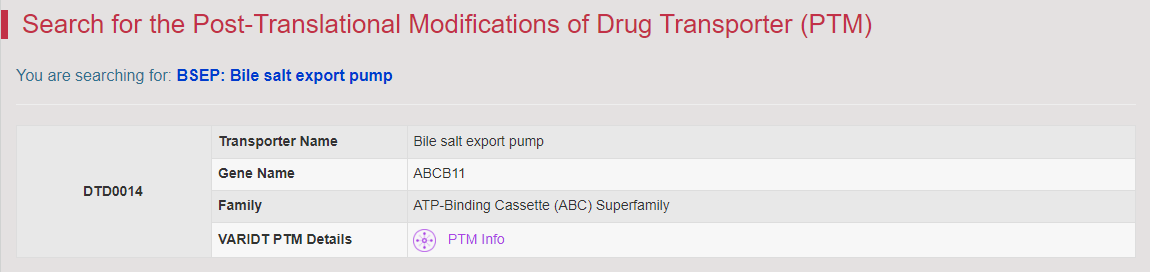
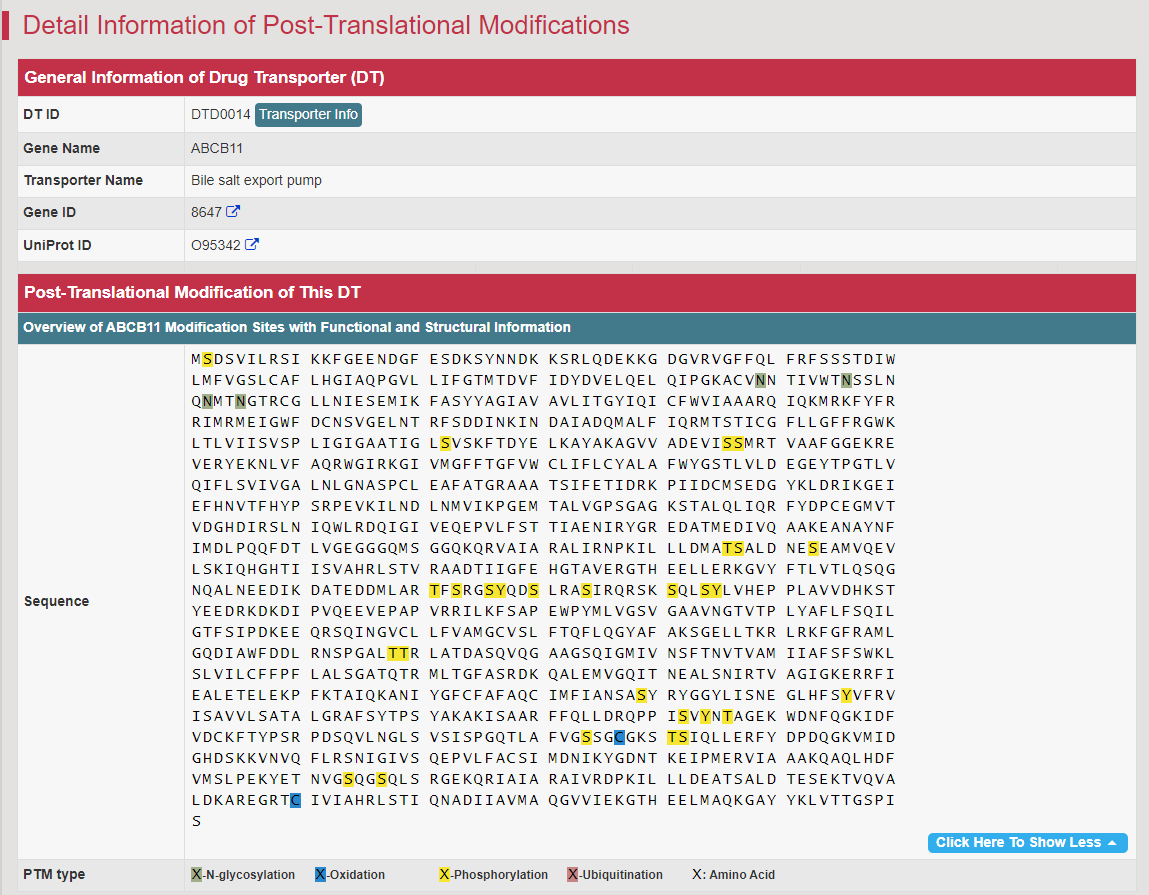
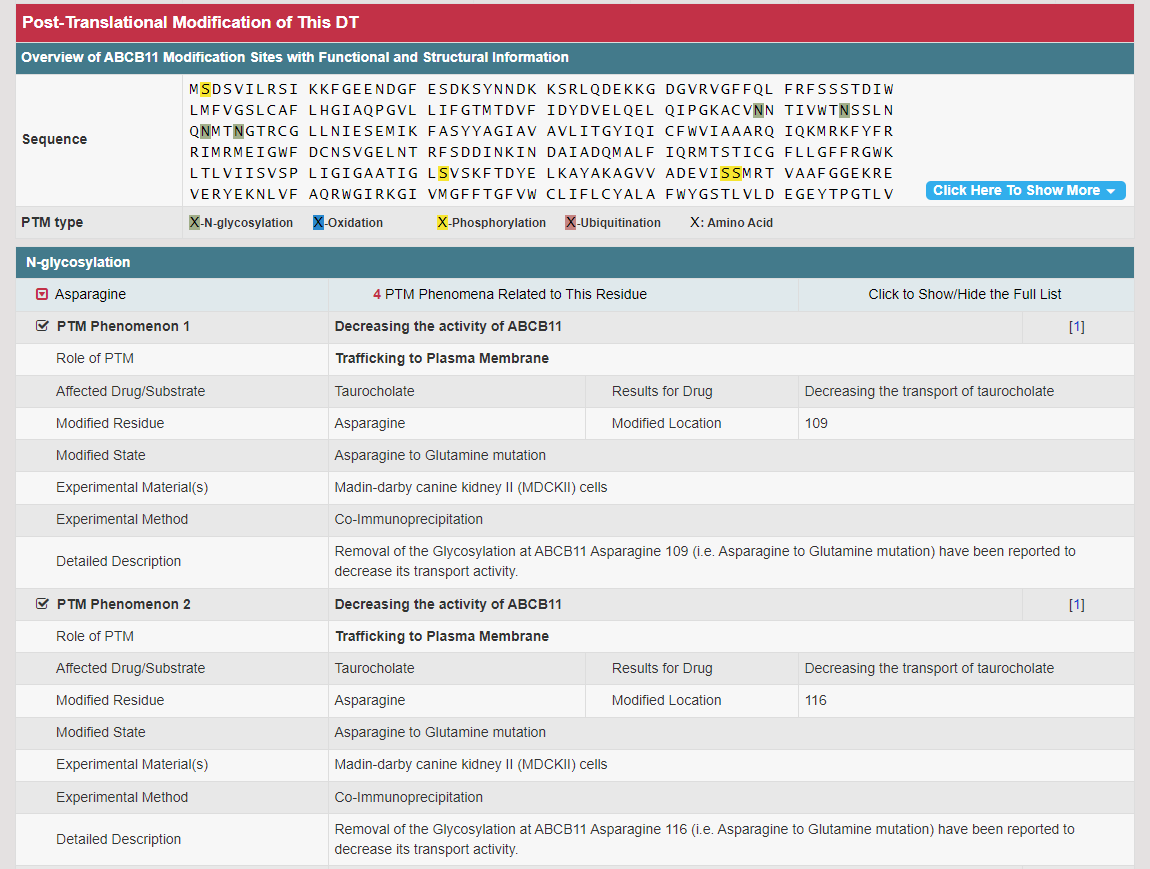
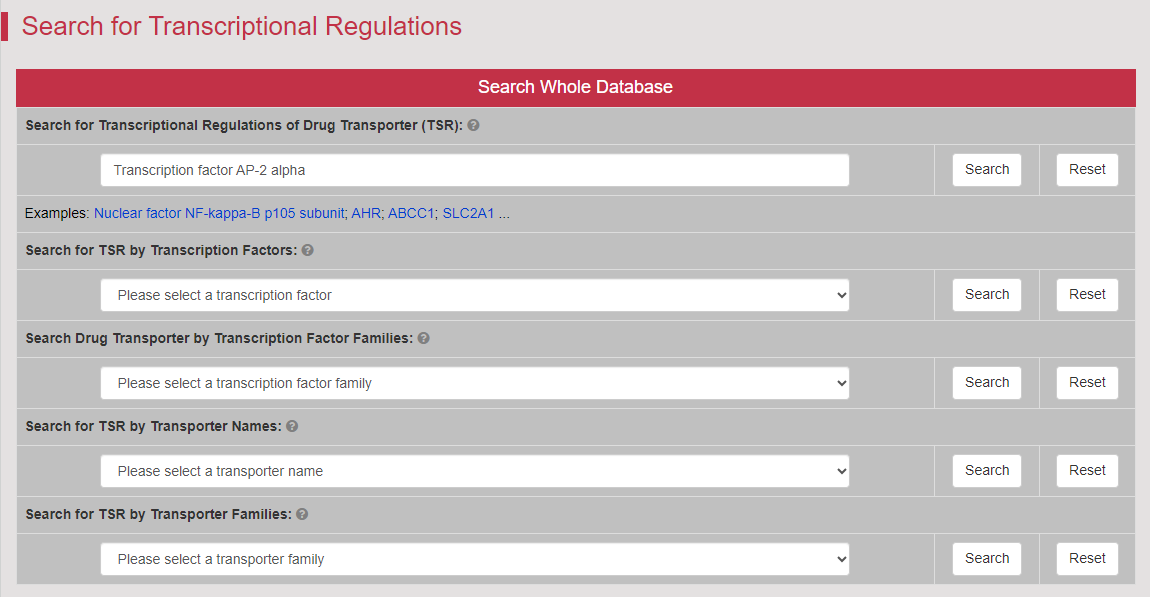
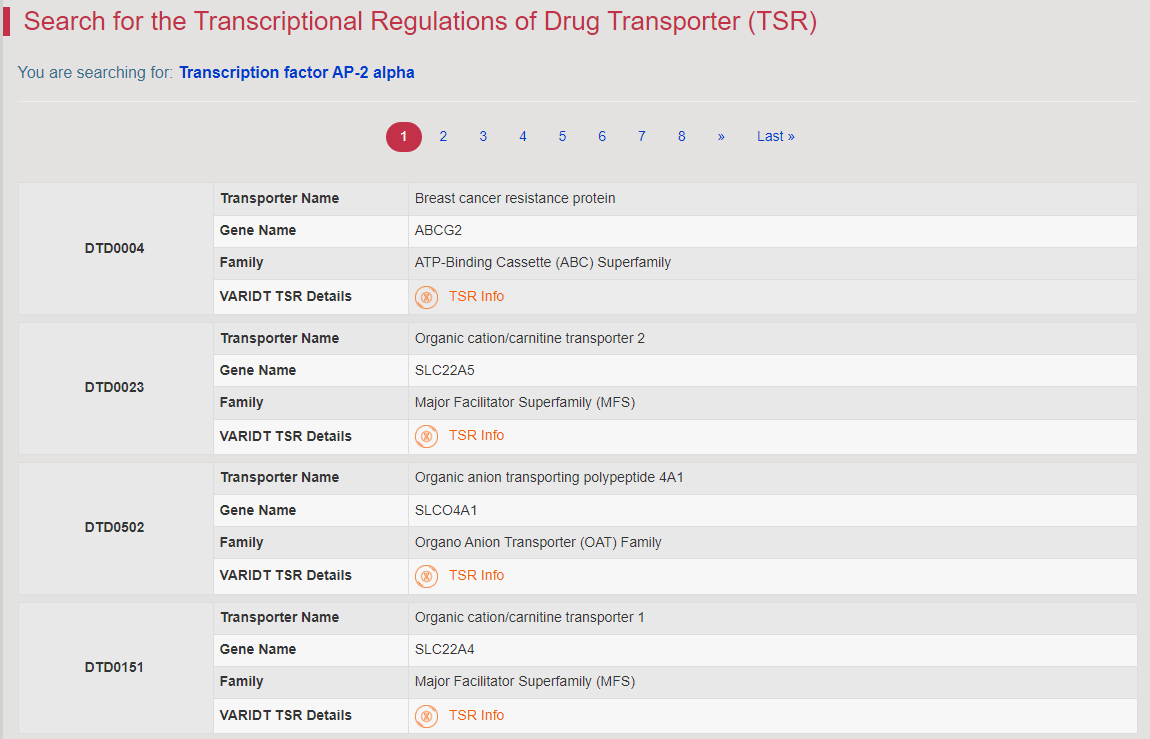
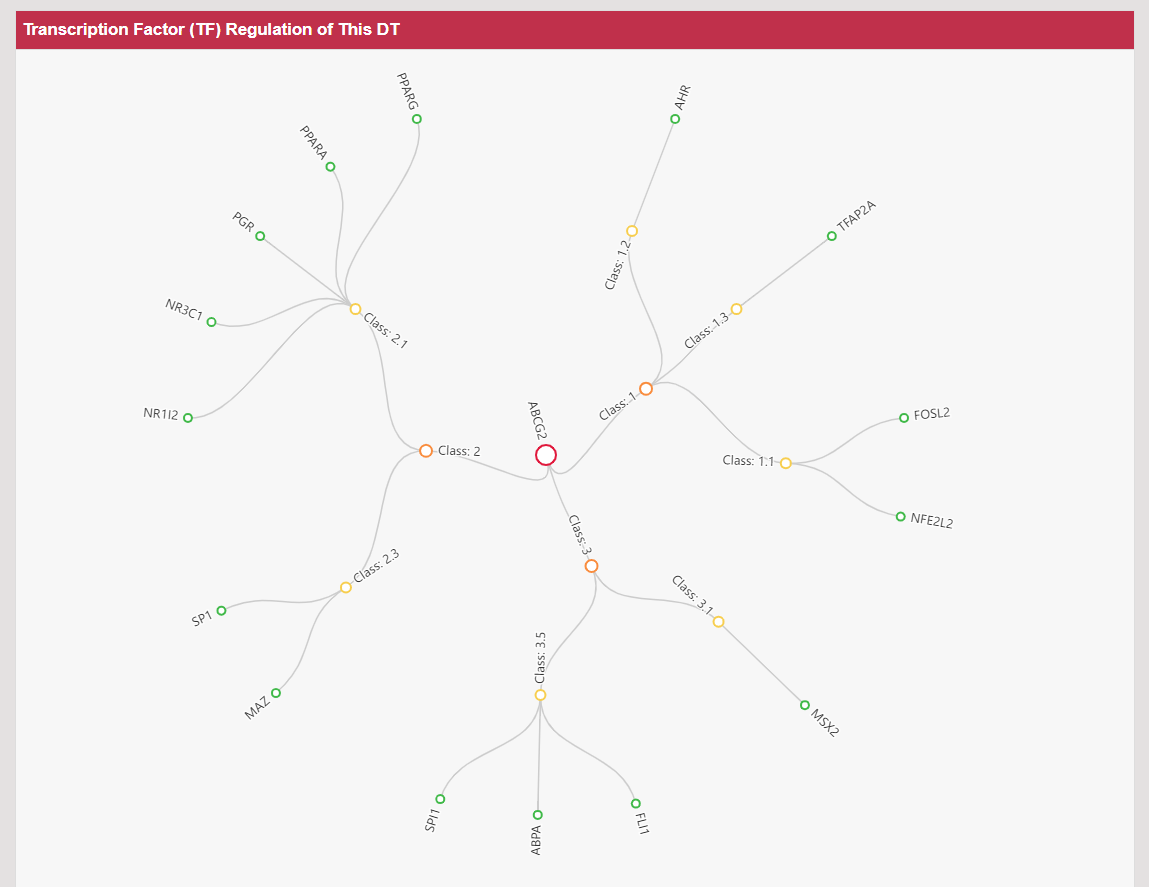
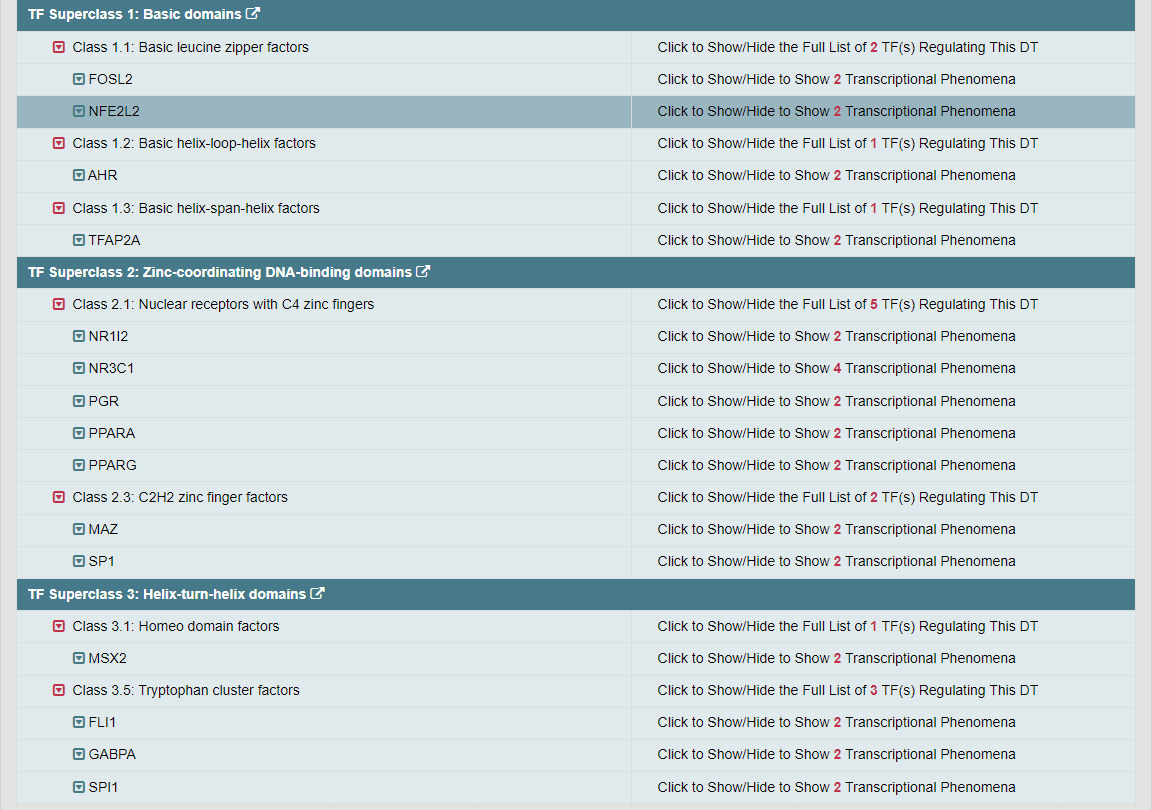
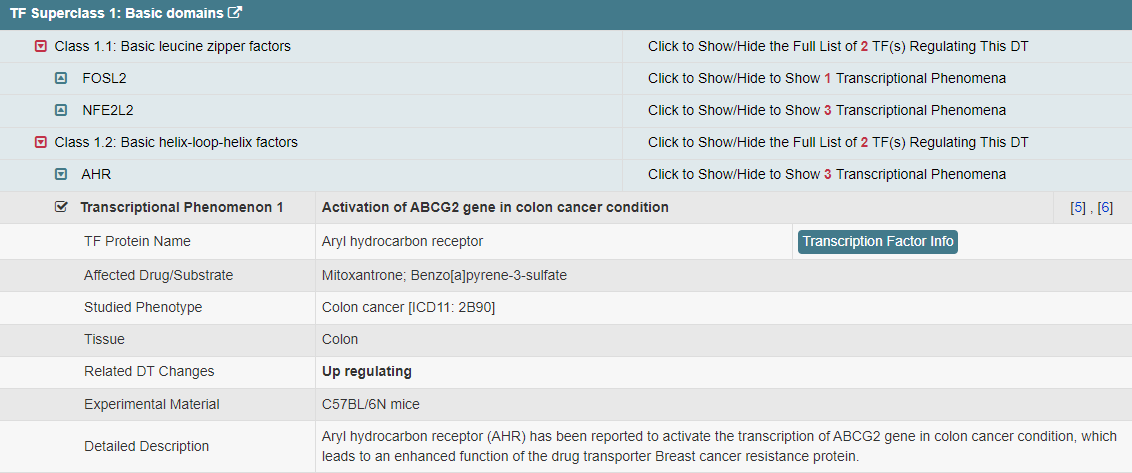
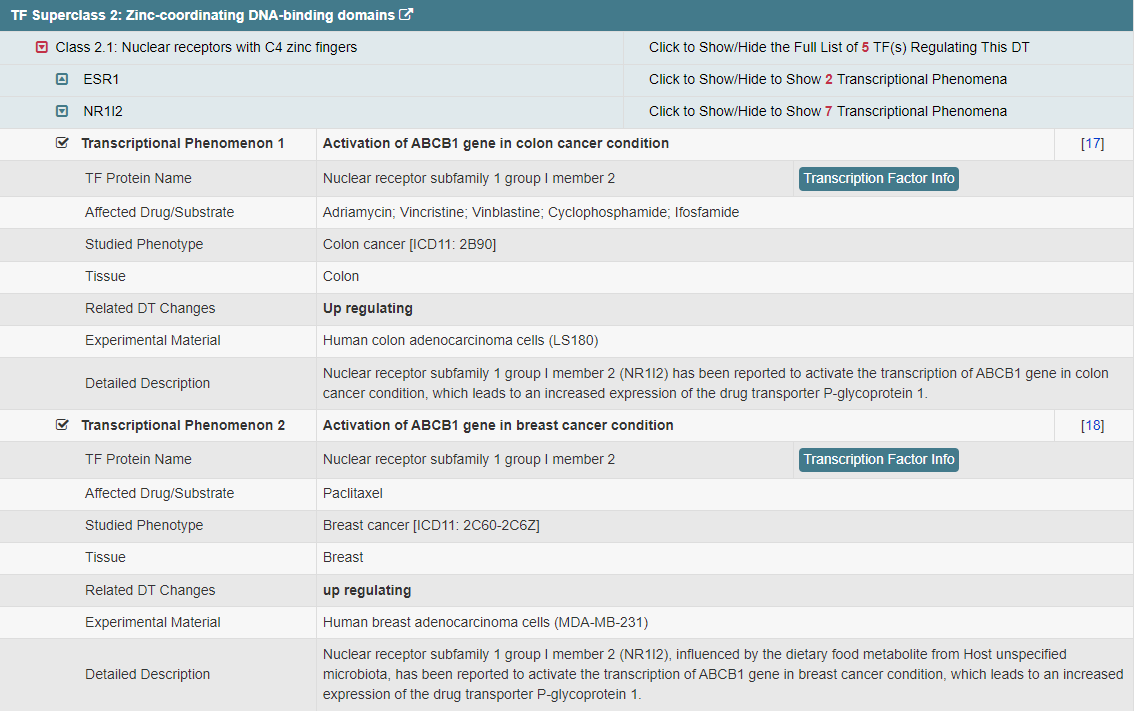
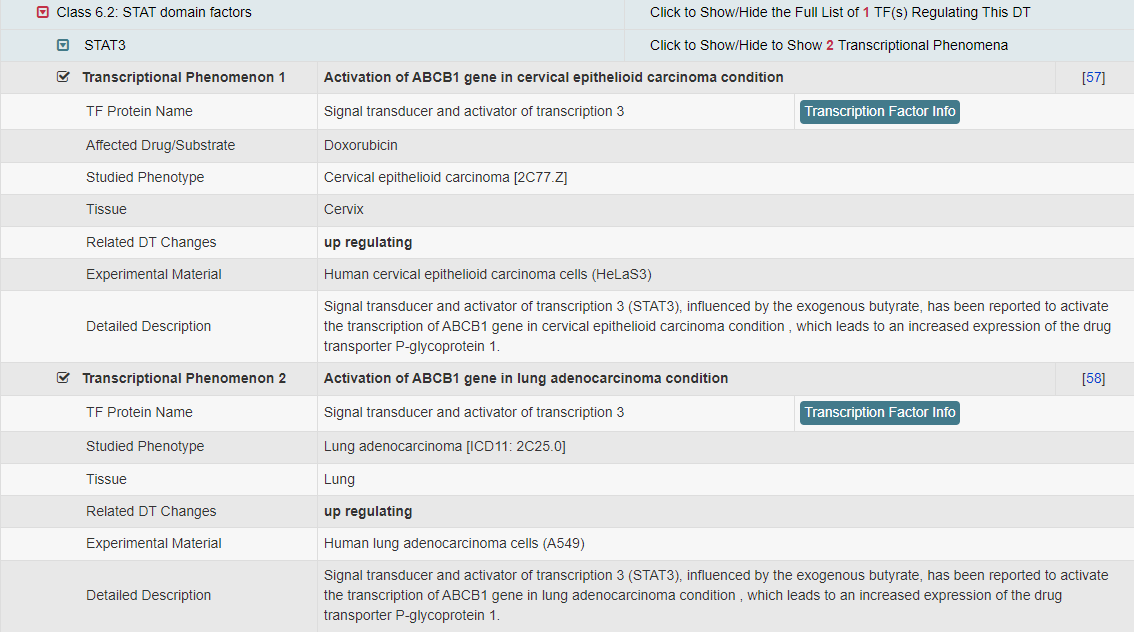
Regulations of drug transporters (DTs) have attracted extensive attentions in recent years, and a variety of regulations have been found to significantly modify the ADME property of a drug by inducing substantial variabilities on the corresponding DT (Pharmacol Ther. 2021, 217: 107647; Physiol Rev. 2022, 102: 993-1024; Clin Pharmacol Ther. 2022, 112: 461-484). A major update of VARIDT was conducted, which systematically described all three aspects of DT regulatory variability: (a) the microbiota influence of DTs, (b) the post-translational modification of DTs, (c) the transcriptional regulation of DTs, (d) the epigenetic regulation of DTs, and (e) the exogenous modulation of DTs. |
Structural Variability (2022 Update)
| Structural Variability (2022 Update) |
|---|
|
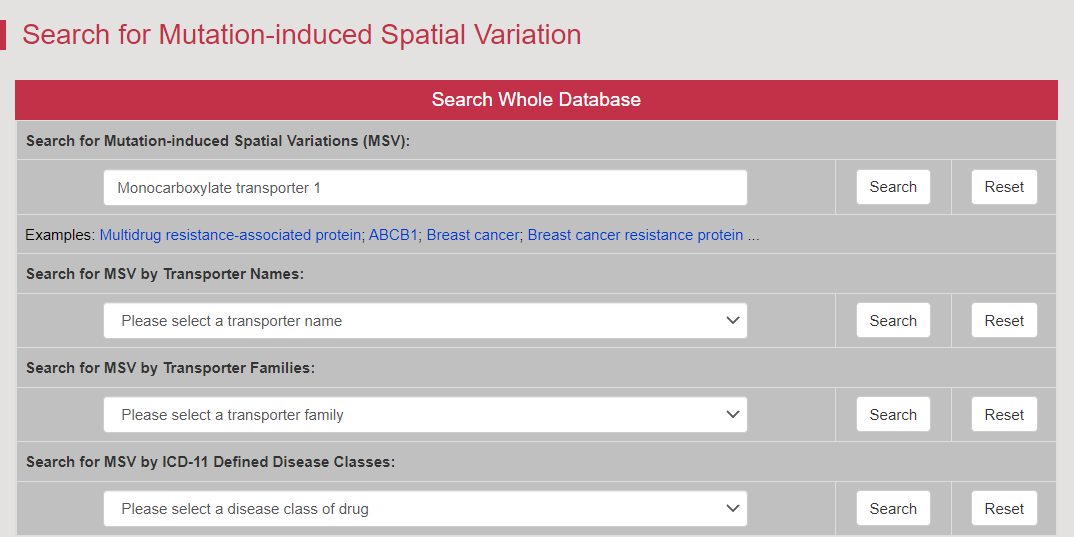
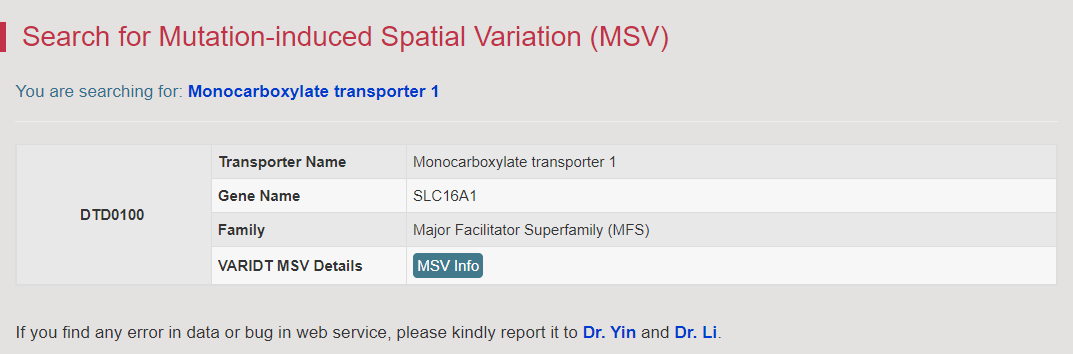
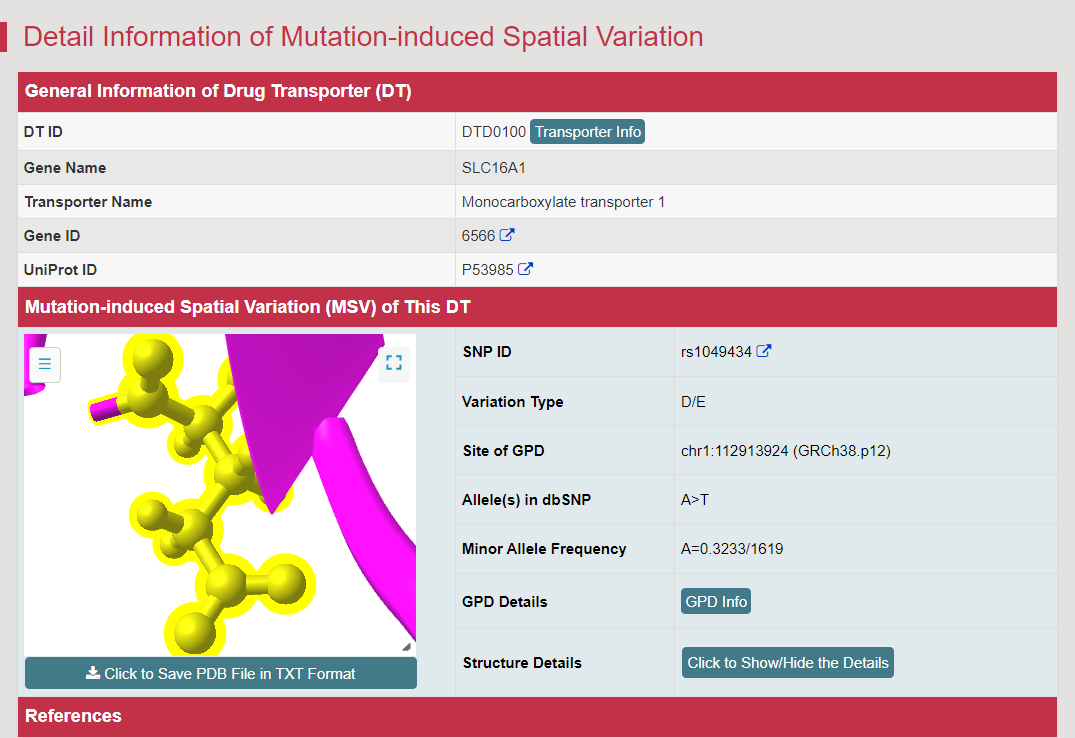
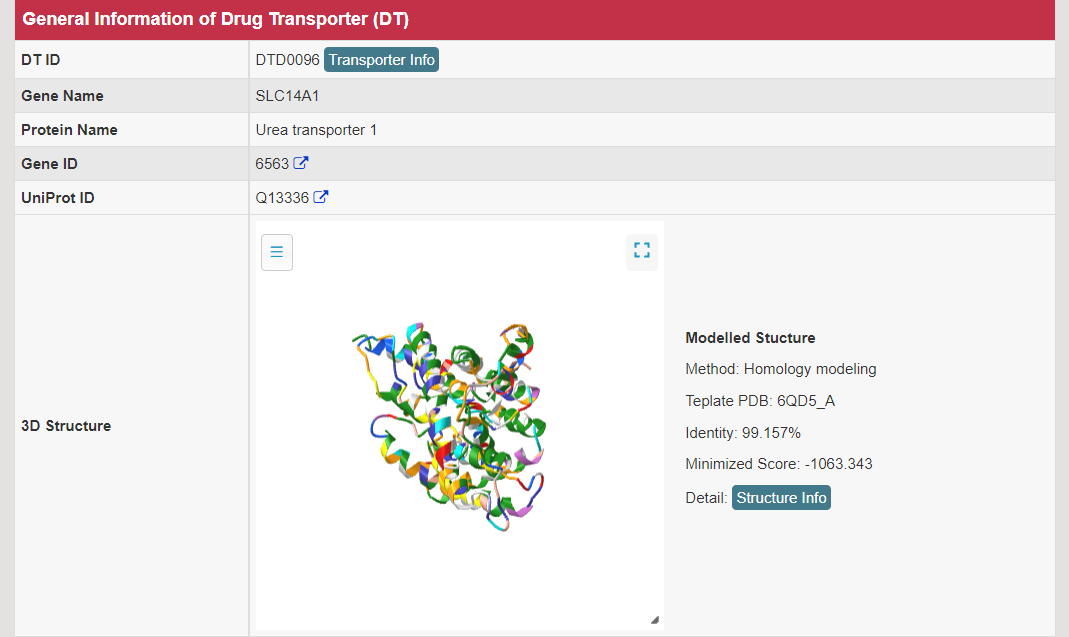
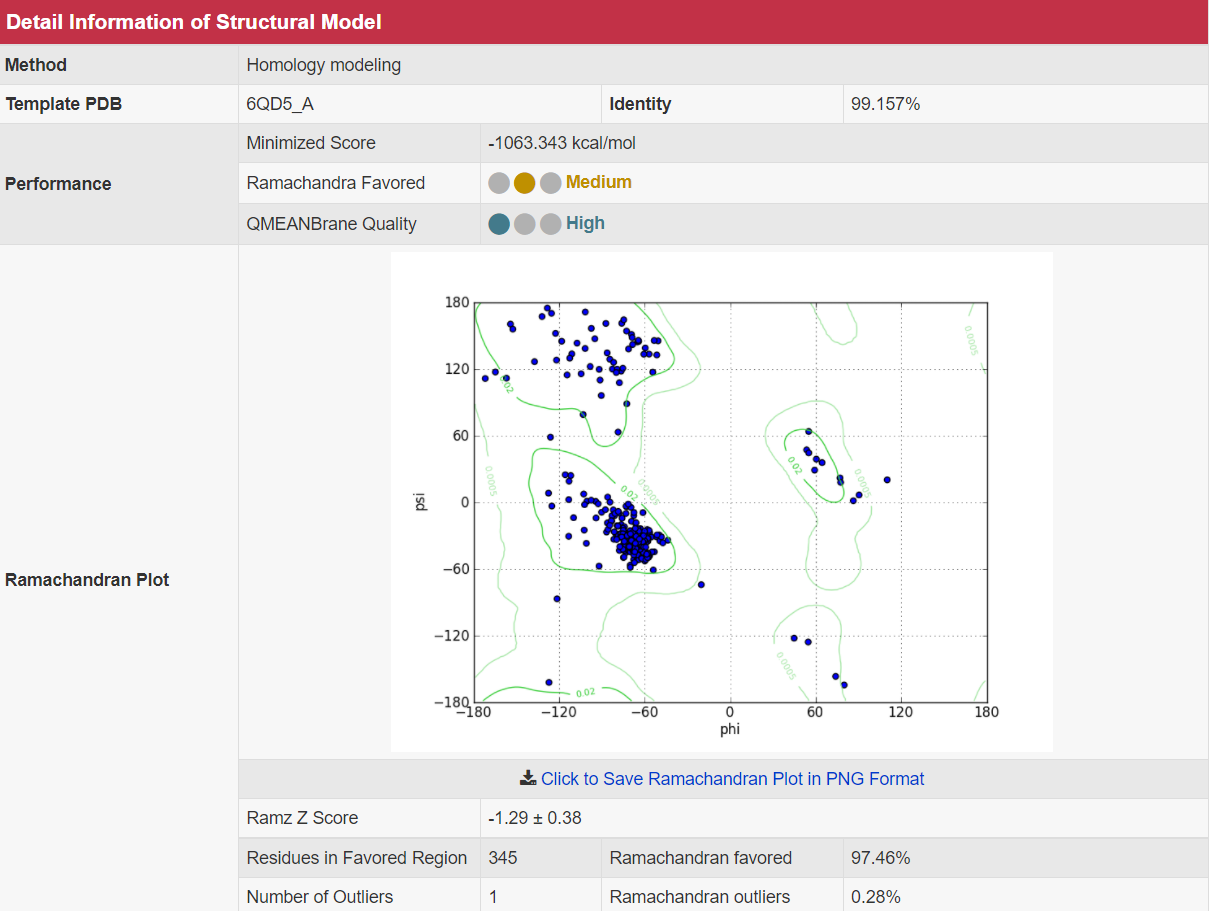
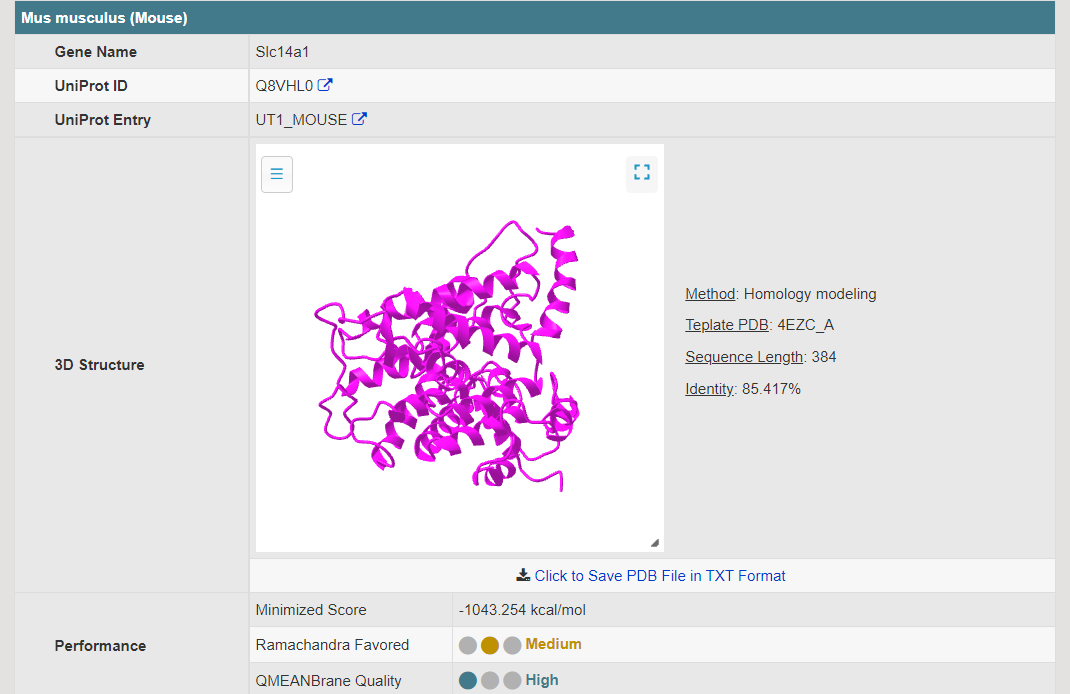
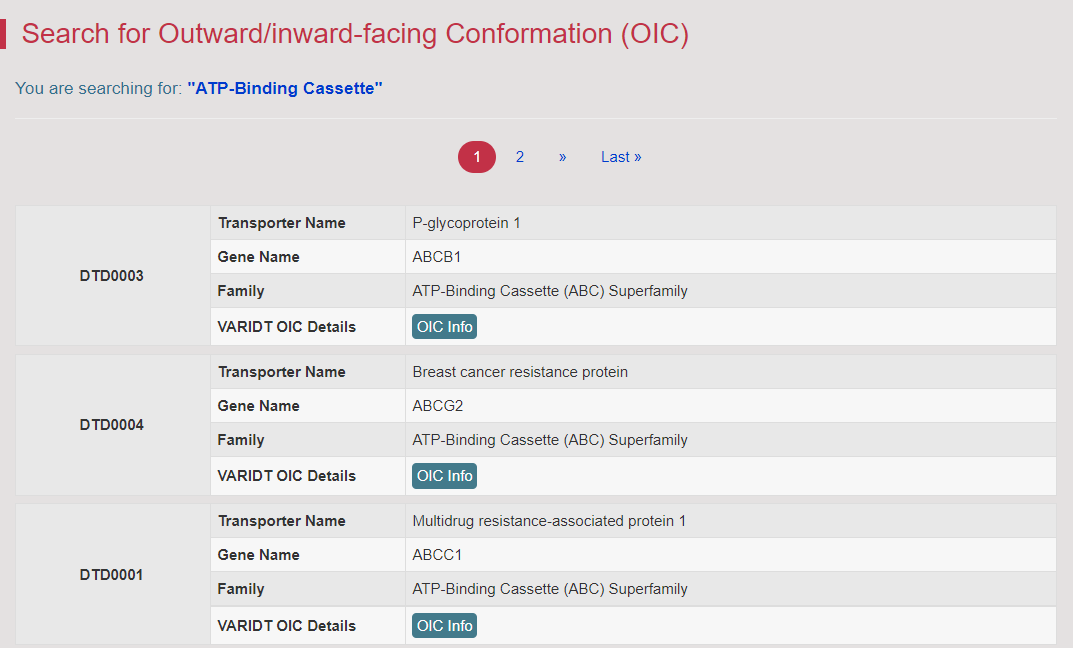
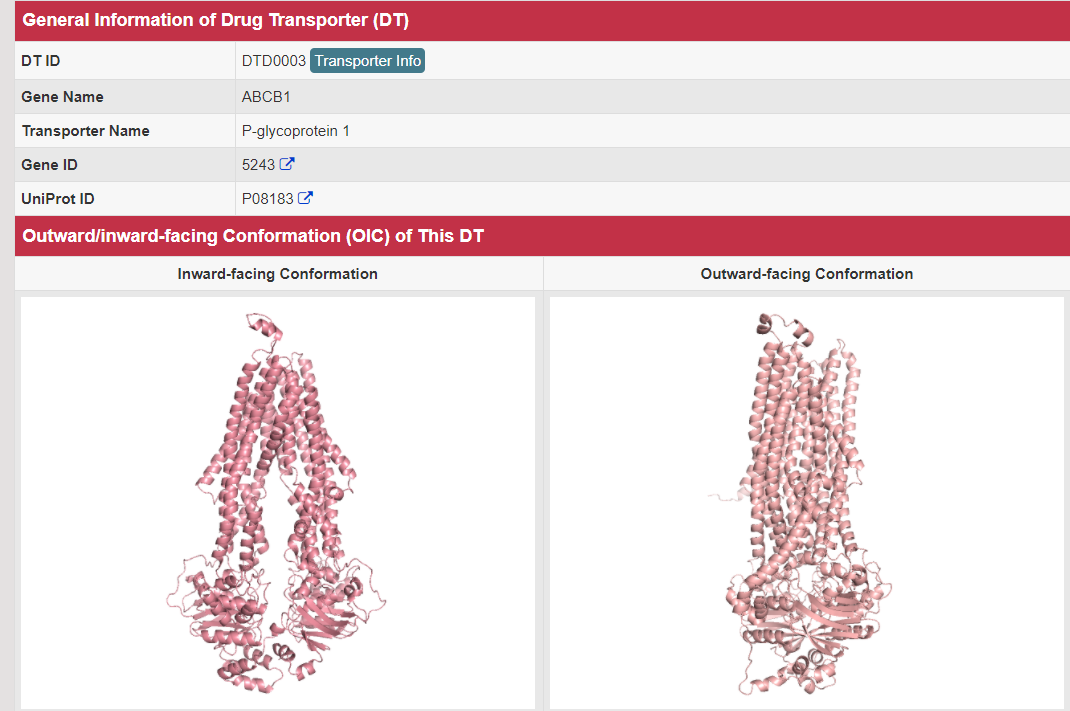
There are increasing demands on the three-dimensional structural variability data of DT, which are expected to provide key information for the studies on precision medicine and rational drug use (Nature. 2019, 576: 315-320). A major update of VARIDT was conducted, which systematically described all four aspects of DT structural variability: (a) the mutation-induced spatial variations in folded state, (b) the difference among the DT structures of human and model organisms, (c) the outward-facing and inward-facing conformations of the transporting cycle, and (d) the xenobiotics-driven alterations in 3D complex. |
| Xenobiotics-driven Alterations in 3D Complexes |
|---|
|
The xenobiotics-driven alterations were essential for revealing mechanisms underlying drug-drug interactions (Elife. 2020, 9: e56427). A total of 822 xenobiotics-regulated structures of 506 xenobiotics in complex with 57 DTs are collected and described in this section. The users can obtain the detail information of xenobiotics-driven alterations in 3D complexes by searching the transporter name (taking SERT as an example) in the “Search for Xenobiotics-driven Alterations in 3D Complexes (XSV):” field. 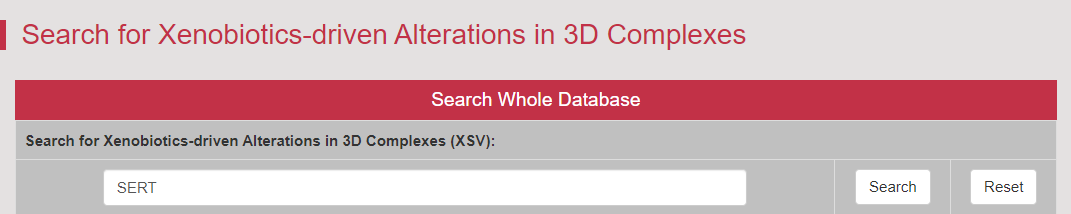
The XSV Info button links to the Detail Information of Xenobiotics-driven Alterations in 3D Complexes page. 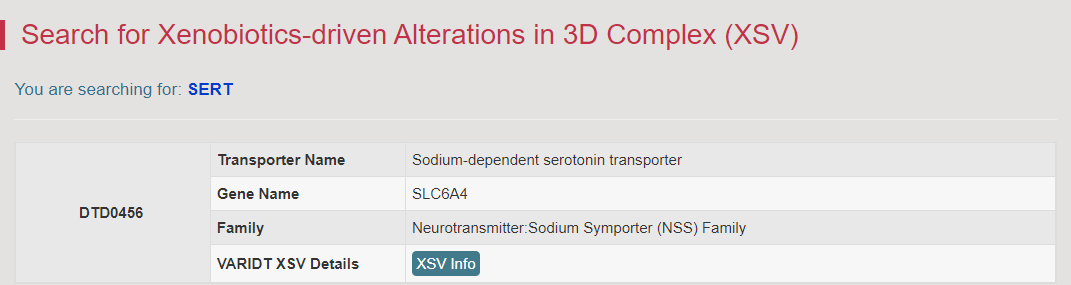
All of the xenobiotics-binding complexes of this DT are provided in this page. Furthermore, more drug information and exogenous factor modulated information can be obtained from the Drug Info and EFMD Info buttons. 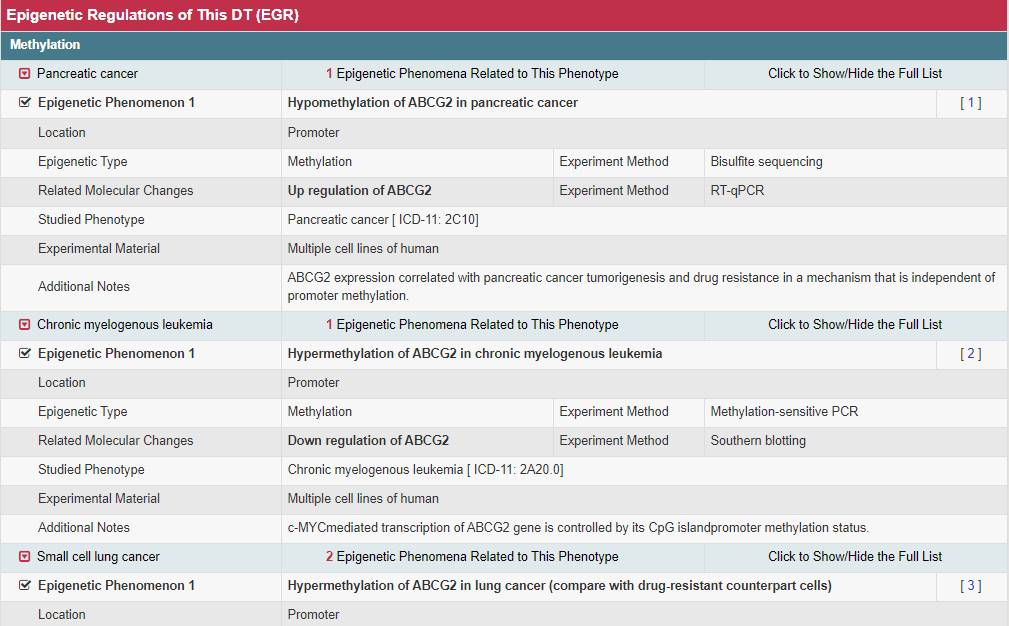
|
Variability of DT
| Variability of DT |
|---|
|
The variability of DT has attracted considerable attentions and broad interests (Nat Rev Drug Discov. 2015, 14: 29-44; Ann Oncol. 2013, 24: 1513-25). This section describes three aspects of DT variability: (a) genetic polymorphism, and epigenetic regulation of DT; (b) species/tissue/disease-specific DT abundance; and (c) exogenous factors modulating DT activity or altering the disposition of transported drug. |
| Genetic/Epigenetic Variability |
|---|
|
Epigenetic regulation and genetic polymorphism of DT that are key in drug resistance and clinical treatment optimization (Sci Transl Med. 2016, 8: 348ra397; Adv Drug Deliv Rev. 2017, 116: 21–36). A total of 38153 DNA methylations, 7317 noncoding RNA/histone regulations, and 1278 genetic polymorphisms are provided. Searching "ABCG2" in the "Search for Drug Transporter Entries:" field, the EGR Info button links to the Detail Information of Epigenetic Regulations page. In the “Epigenetic Regulations of DT (EGR)” section, the epigenetic regulations information including three aspects “methylation”, “histone acetylation” and “microRNA” are provided. 
Moreover, the detail information of each epigenetic regulation (methylation, histone acetylation and microRNA) is displayed by a list. 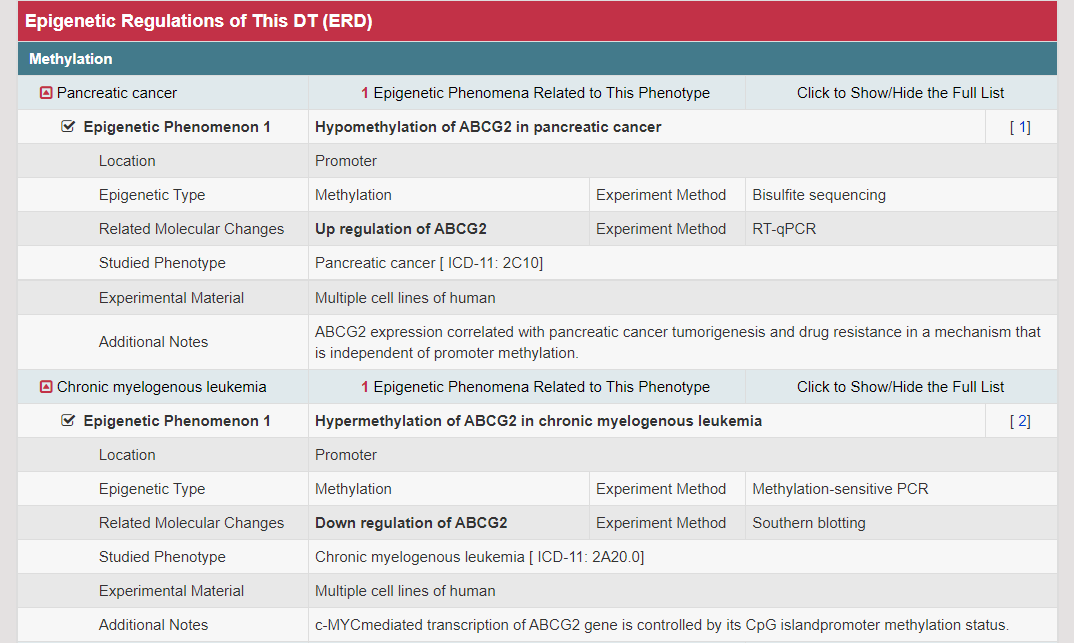
|
| Protein Abundance Variability |
|---|
|
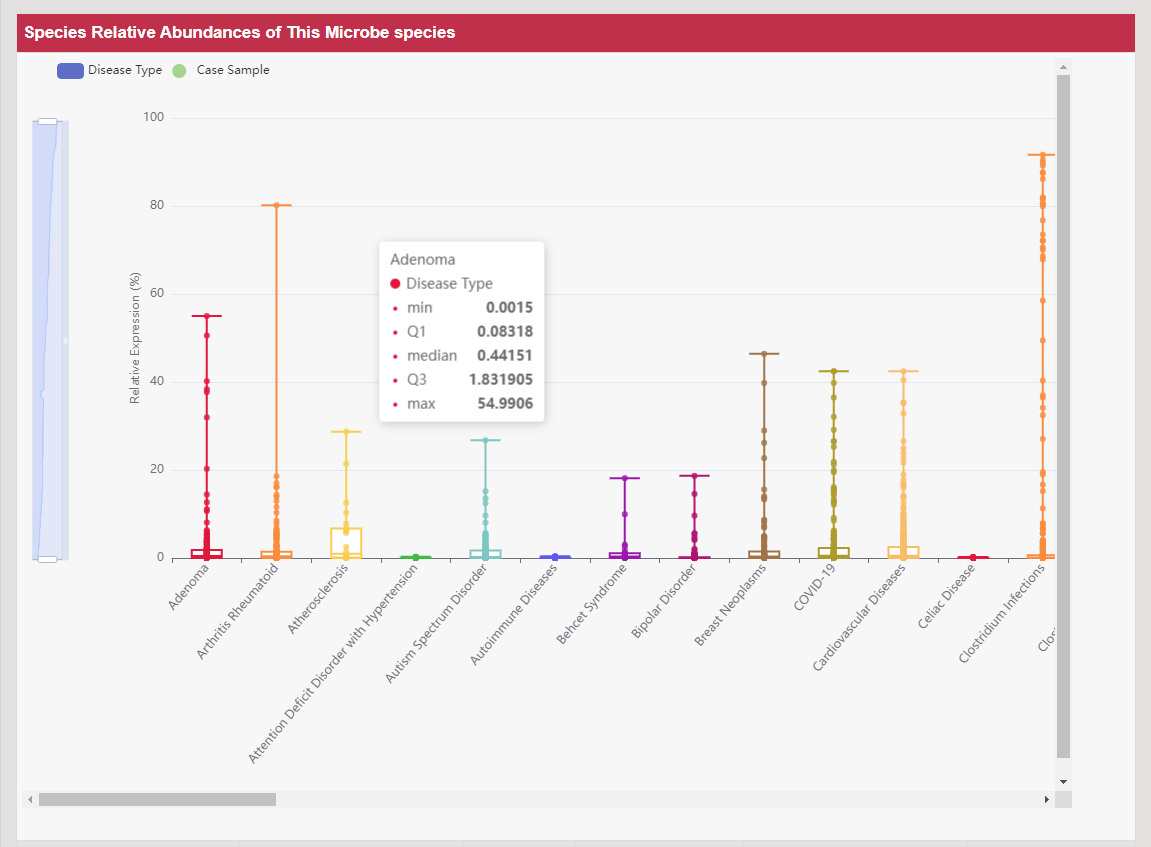
Protein abundance of DT was always a critical issue in various aspects of drug development, such as clinical pharmacokinetics, adverse reaction assessments and drug-drug interactions (Nat Rev Drug Discov. 2015, 14: 543–560). Differential abundance profiles of 257 DTs in 21 781 patients/healthy individuals, expression of 245 DTs in 67 tissues of human/model organism are shown in this section. Taking ATP-binding cassette sub-family A member 1 as an example, searching the “ABCA1: ATP-binding cassette sub-family A member 1” in the “Search for DPAD by Transporter Names:” field. The Detail Information of Disease-specific Protein Abundances page can be accessed by clicking the DPAD Info button. Multiple aspects of data are presented by a list. 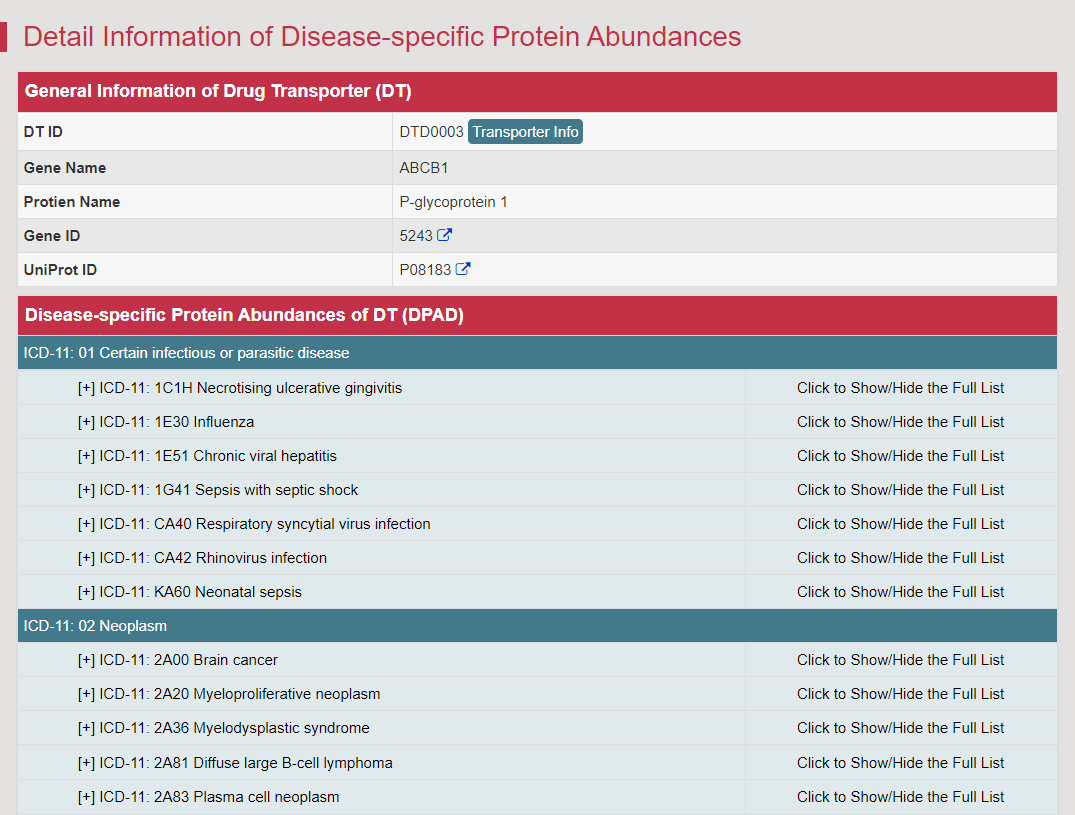
|
| Exogenous Modulation |
|---|
|
The activity or expression of DTs could be inhibited or induced by exogenous factors, which, in turn, affected the pharmacokinetics, efficacy and safety of drugs or the tissue level of drugs/substrates that were transported by the corresponding DTs (Nat Rev Drug Discov. 2010, 9: 215–236). A total of 1717 exogenous factors altering the activity of 419 DTs are collected in this section. For example, searching the “Ketoconazole” in the “Search for Exogenous Modulation of Drug Transporter (EGM)” filed. The result displays several related DTs. 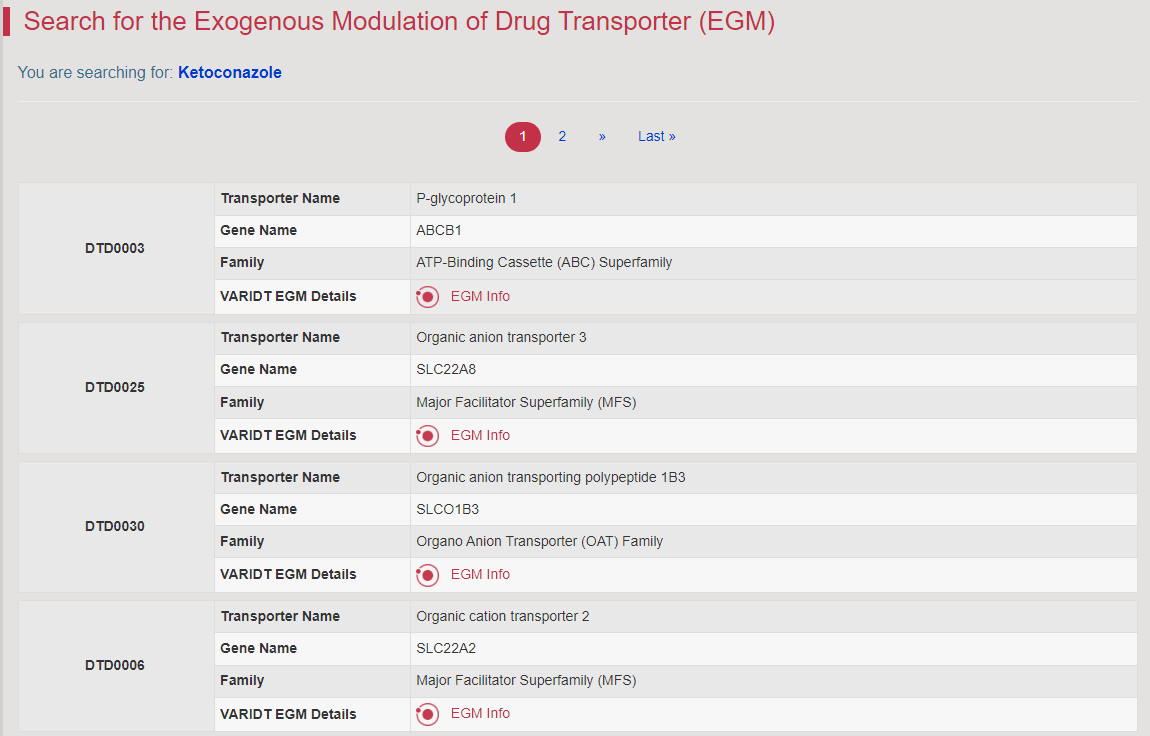
The Information of Exogenous Factor Modulation page can be accessed by clicking the EGM Info button. This page provides various detail information of the exogenous factors (drugs have been approved or in in clinical/preclinical studies, natural product, dietary constituent, environmental toxicant, pharmaceutical excipient, etc.), related drug/substrate, modulation type and activity. 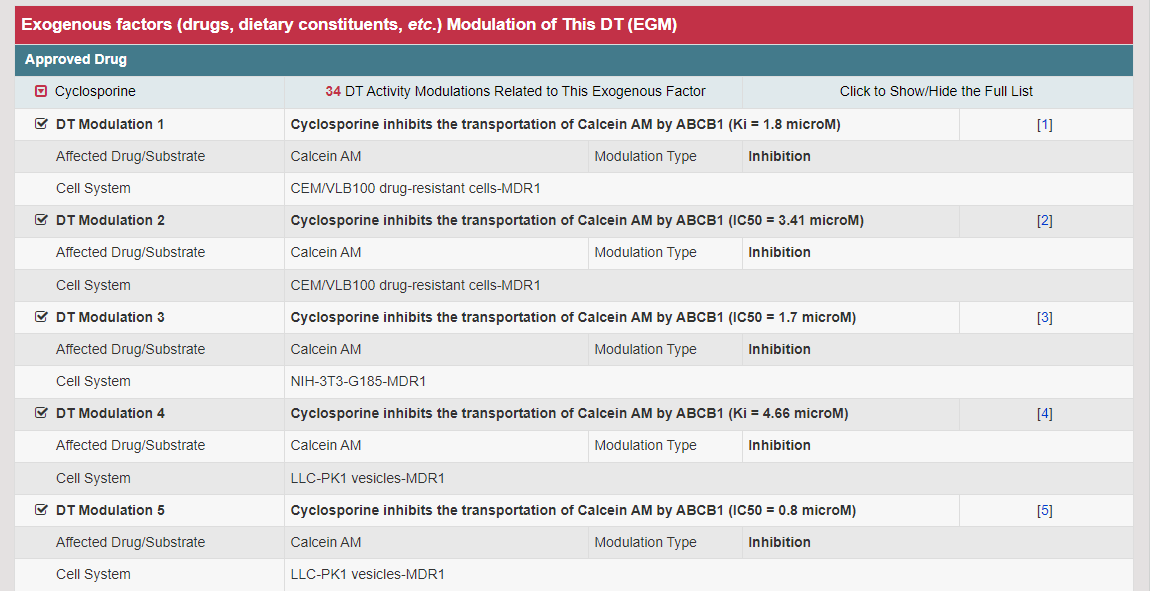
It should be noted that the impact of EGM on DT may be achieved through certain specific regulatory mechanisms. In VARIDT, the specific mechanisms of the impact of literature on reported EGM are included and provided on the webpage, highlighted in bold. Users can also click the corresponding button to jump to the specific page of the regulatory mechanism. 
|
Download
| Download |
|---|
|
Download the Full Data of VARIDT from a Variety of Customized Links. VARIDT provides functions for downloading all VARIDR data from various customized links. (1) full 3D variability data of DT (mutation-induced spatial variations of DTs, inter species structural differences, outward/inward-facing conformations, xenobiotics-driven alterations in 3D complexes) (2) full DT variability data (epigenetic regulation data of DTs, genetic polymorphism data of DTs, disease-specific protein abundances of DTs, species-specific protein abundances of DTs, tissue-specific protein abundances of DTs and exogenous factors altering DT activity) (3) full sequence/structure/function data/ cross-matching data (sequence and structure data of DTs, transporter-drug affinity data based on cell line experiment, functional family data for each DT, 2D structure data for the drugs transported by DTs, 3D structure data for the drugs transported by DTs, SMILES and InChI for the drugs transported by DTs and therapeutic classes and physicochemical properties for the drugs, cross-matching ID between the DTs and a variety of public databases, synonyms of DTs and their corresponding drugs, DTs to disease mapping with ICD identifiers and Drug to disease mapping with ICD-11 identifiers) All data can be readily downloaded by simply clicking the corresponding 'Click to Save' button. |