Detail Information of Epigenetic Regulations
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0006 Transporter Info | ||||
| Gene Name | SLC22A2 | ||||
| Transporter Name | Organic cation transporter 2 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| Epigenetic Regulations of This DT (EGR) | |||||
|---|---|---|---|---|---|
|
Methylation |
|||||
|
Renal cell carcinoma |
4 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Hypermethylation of SLC22A2 in renal cell carcinoma | [ 1 ] | |||
|
Location |
Proximal promoter | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Bisulfite sequencing | ||
|
Related Molecular Changes |
Down regulation of SLC22A2 | Experiment Method | RT-qPCR | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Frequency |
70% of the studied tumor samples | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Additional Notes |
All CpG sites in the OCT2 proximal promoter were hypomethylated in the kidney. | ||||
|
Epigenetic Phenomenon 2 |
Hypermethylation of SLC22A2 in renal cell carcinoma | [ 2 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Bisulfite sequencing PCR | ||
|
Related Molecular Changes |
Down regulation of SLC22A2 | Experiment Method | RT-qPCR | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Frequency |
In all three RCC lines (786-O, 769-P, and Caki-1) | ||||
|
Experimental Material |
Multiple cell lines of human; Patients tissue samples | ||||
|
Additional Notes |
DNA Hypermethylation blocked MYC activation of OCT2 by disrupting its interaction with the E-Box motif, which prevented MYC from recruiting MLL1 to catalyze H3K4me3 at the OCT2 promoter and resulted in repressed OCT2 transcription. | ||||
|
Epigenetic Phenomenon 3 |
Hypermethylation of SLC22A2 in renal cell carcinoma | [ 5 ] | |||
|
Location |
Promoter (10 CpG sites) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | HumanMethylation450 BeadChip | ||
|
Related Molecular Changes |
Down regulation of SLC22A2 | Experiment Method | Affymetrix Human Transcriptome 2.0 microarrays | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
In RCC cell lines, OCT2 protein expression is downregulated due to Hypermethylation in the SLC22A2 promoter region compared to primary tumors or metastases. | ||||
|
Esophageal cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Hypermethylation of SLC22A2 in cisplatin-resistant esophageal cancer (compare with parental cells) | [ 3 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Methylation-sensitive PCR | ||
|
Related Molecular Changes |
Down regulation of SLC22A2 | Experiment Method | Western Blot | ||
|
Studied Phenotype |
Esophageal cancer [ ICD-11: 2B70] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Additional Notes |
Long-term exposure to cisplatin promotes methylation of the OCT1 gene in human esophageal cancer cells. | ||||
|
Hepatocellular carcinoma |
12 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Hypermethylation of SLC22A2 in hepatocellular carcinoma (compare with non-tumor adjacent tissues) | [ 4 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Methylation | Experiment Method | MALDI-TOF MS | ||
|
Related Molecular Changes |
Down regulation of SLC22A2 | Experiment Method | RT-qPCR | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Additional Notes |
DNA methylation is associated with downregulation of the organic cation transporter OCT1 (SLC22A1) in human hepatocellular carcinoma. | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC22A2 in hepatocellular carcinoma | [ 12 ] | |||
|
Location |
TSS1500 (cg07026448) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.16E+00 | Statistic Test | p-value: 1.17E-08; Z-score: 1.41E+00 | ||
|
Methylation in Case |
6.40E-01 (Median) | Methylation in Control | 5.50E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC22A2 in hepatocellular carcinoma | [ 12 ] | |||
|
Location |
TSS1500 (cg23995605) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.01E+00 | Statistic Test | p-value: 8.40E-03; Z-score: -3.12E-01 | ||
|
Methylation in Case |
8.79E-01 (Median) | Methylation in Control | 8.87E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC22A2 in hepatocellular carcinoma | [ 12 ] | |||
|
Location |
TSS1500 (cg14450766) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.07E+00 | Statistic Test | p-value: 3.95E-02; Z-score: 3.63E-01 | ||
|
Methylation in Case |
4.92E-01 (Median) | Methylation in Control | 4.61E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC22A2 in hepatocellular carcinoma | [ 12 ] | |||
|
Location |
TSS200 (cg06236276) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.04E+00 | Statistic Test | p-value: 8.34E-03; Z-score: 1.72E-01 | ||
|
Methylation in Case |
6.85E-01 (Median) | Methylation in Control | 6.60E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC22A2 in hepatocellular carcinoma | [ 12 ] | |||
|
Location |
Body (cg10473100) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.24E+00 | Statistic Test | p-value: 6.30E-12; Z-score: -1.29E+01 | ||
|
Methylation in Case |
7.82E-01 (Median) | Methylation in Control | 9.73E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC22A2 in hepatocellular carcinoma | [ 12 ] | |||
|
Location |
Body (cg08793936) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.26E+00 | Statistic Test | p-value: 1.84E-10; Z-score: -3.25E+00 | ||
|
Methylation in Case |
5.62E-01 (Median) | Methylation in Control | 7.09E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC22A2 in hepatocellular carcinoma | [ 12 ] | |||
|
Location |
Body (cg04310488) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.46E+00 | Statistic Test | p-value: 1.36E-06; Z-score: -1.32E+00 | ||
|
Methylation in Case |
2.63E-01 (Median) | Methylation in Control | 3.84E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLC22A2 in hepatocellular carcinoma | [ 12 ] | |||
|
Location |
Body (cg14918008) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.02E+00 | Statistic Test | p-value: 3.70E-03; Z-score: -7.65E-01 | ||
|
Methylation in Case |
8.01E-01 (Median) | Methylation in Control | 8.19E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 10 |
Methylation of SLC22A2 in hepatocellular carcinoma | [ 12 ] | |||
|
Location |
Body (cg17098387) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.07E+00 | Statistic Test | p-value: 4.77E-03; Z-score: 4.43E-01 | ||
|
Methylation in Case |
7.81E-01 (Median) | Methylation in Control | 7.33E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 11 |
Methylation of SLC22A2 in hepatocellular carcinoma | [ 12 ] | |||
|
Location |
Body (cg07601258) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.01E+00 | Statistic Test | p-value: 1.45E-02; Z-score: -3.44E-01 | ||
|
Methylation in Case |
8.71E-01 (Median) | Methylation in Control | 8.80E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 12 |
Methylation of SLC22A2 in hepatocellular carcinoma | [ 12 ] | |||
|
Location |
3'UTR (cg04092332) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.16E+00 | Statistic Test | p-value: 5.29E-11; Z-score: -4.69E+00 | ||
|
Methylation in Case |
7.42E-01 (Median) | Methylation in Control | 8.63E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Prostate cancer |
20 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC22A2 in prostate cancer | [ 6 ] | |||
|
Location |
5'UTR (cg13323097) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.51E+00 | Statistic Test | p-value: 1.89E-04; Z-score: 5.73E+00 | ||
|
Methylation in Case |
2.20E-01 (Median) | Methylation in Control | 1.46E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC22A2 in prostate cancer | [ 6 ] | |||
|
Location |
5'UTR (cg26248082) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.19E+00 | Statistic Test | p-value: 3.00E-02; Z-score: 2.67E+00 | ||
|
Methylation in Case |
7.67E-01 (Median) | Methylation in Control | 6.44E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC22A2 in prostate cancer | [ 6 ] | |||
|
Location |
TSS1500 (cg01352551) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.11E+00 | Statistic Test | p-value: 3.97E-03; Z-score: 2.49E+00 | ||
|
Methylation in Case |
8.12E-01 (Median) | Methylation in Control | 7.34E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC22A2 in prostate cancer | [ 6 ] | |||
|
Location |
TSS1500 (cg17174275) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.09E+00 | Statistic Test | p-value: 9.95E-03; Z-score: 2.19E+00 | ||
|
Methylation in Case |
8.67E-01 (Median) | Methylation in Control | 7.96E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC22A2 in prostate cancer | [ 6 ] | |||
|
Location |
TSS1500 (cg27043188) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.45E+00 | Statistic Test | p-value: 1.29E-02; Z-score: -2.00E+00 | ||
|
Methylation in Case |
3.22E-01 (Median) | Methylation in Control | 4.66E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC22A2 in prostate cancer | [ 6 ] | |||
|
Location |
TSS1500 (cg10528482) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.21E+00 | Statistic Test | p-value: 1.51E-02; Z-score: 1.57E+01 | ||
|
Methylation in Case |
8.28E-01 (Median) | Methylation in Control | 6.86E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC22A2 in prostate cancer | [ 6 ] | |||
|
Location |
TSS1500 (cg04833514) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.17E+00 | Statistic Test | p-value: 1.91E-02; Z-score: 3.12E+00 | ||
|
Methylation in Case |
8.25E-01 (Median) | Methylation in Control | 7.03E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC22A2 in prostate cancer | [ 6 ] | |||
|
Location |
TSS1500 (cg02707152) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.44E+00 | Statistic Test | p-value: 3.01E-02; Z-score: 3.38E+00 | ||
|
Methylation in Case |
3.34E-01 (Median) | Methylation in Control | 2.31E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLC22A2 in prostate cancer | [ 6 ] | |||
|
Location |
TSS200 (cg18335740) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.62E+00 | Statistic Test | p-value: 3.86E-03; Z-score: 3.84E+00 | ||
|
Methylation in Case |
2.91E-01 (Median) | Methylation in Control | 1.80E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 10 |
Methylation of SLC22A2 in prostate cancer | [ 6 ] | |||
|
Location |
1stExon (cg08457872) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.05E+00 | Statistic Test | p-value: 3.39E-02; Z-score: 1.73E+00 | ||
|
Methylation in Case |
9.01E-01 (Median) | Methylation in Control | 8.56E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 11 |
Methylation of SLC22A2 in prostate cancer | [ 6 ] | |||
|
Location |
Body (cg06635952) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 3.11E+00 | Statistic Test | p-value: 4.23E-03; Z-score: 1.29E+01 | ||
|
Methylation in Case |
6.50E-01 (Median) | Methylation in Control | 2.09E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 12 |
Methylation of SLC22A2 in prostate cancer | [ 6 ] | |||
|
Location |
Body (cg06145736) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.21E+00 | Statistic Test | p-value: 6.78E-03; Z-score: 2.70E+00 | ||
|
Methylation in Case |
8.80E-01 (Median) | Methylation in Control | 7.29E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 13 |
Methylation of SLC22A2 in prostate cancer | [ 6 ] | |||
|
Location |
Body (cg25575590) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.39E+00 | Statistic Test | p-value: 8.54E-03; Z-score: -1.19E+01 | ||
|
Methylation in Case |
6.48E-01 (Median) | Methylation in Control | 9.02E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 14 |
Methylation of SLC22A2 in prostate cancer | [ 6 ] | |||
|
Location |
Body (cg15930240) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.33E+00 | Statistic Test | p-value: 1.08E-02; Z-score: 1.96E+00 | ||
|
Methylation in Case |
7.95E-01 (Median) | Methylation in Control | 5.98E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 15 |
Methylation of SLC22A2 in prostate cancer | [ 6 ] | |||
|
Location |
Body (cg00551954) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.05E+00 | Statistic Test | p-value: 1.29E-02; Z-score: 2.04E+00 | ||
|
Methylation in Case |
9.33E-01 (Median) | Methylation in Control | 8.85E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 16 |
Methylation of SLC22A2 in prostate cancer | [ 6 ] | |||
|
Location |
Body (cg10665321) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.45E+00 | Statistic Test | p-value: 2.95E-02; Z-score: 2.02E+00 | ||
|
Methylation in Case |
6.35E-01 (Median) | Methylation in Control | 4.38E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 17 |
Methylation of SLC22A2 in prostate cancer | [ 6 ] | |||
|
Location |
Body (cg22931795) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.51E+00 | Statistic Test | p-value: 4.02E-02; Z-score: -2.37E+00 | ||
|
Methylation in Case |
4.12E-01 (Median) | Methylation in Control | 6.20E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 18 |
Methylation of SLC22A2 in prostate cancer | [ 6 ] | |||
|
Location |
Body (cg04954559) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.79E+00 | Statistic Test | p-value: 4.95E-02; Z-score: 4.75E+00 | ||
|
Methylation in Case |
1.41E-01 (Median) | Methylation in Control | 7.88E-02 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 19 |
Methylation of SLC22A2 in prostate cancer | [ 6 ] | |||
|
Location |
3'UTR (cg09888840) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.06E+00 | Statistic Test | p-value: 8.80E-03; Z-score: 3.47E+00 | ||
|
Methylation in Case |
9.35E-01 (Median) | Methylation in Control | 8.81E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Bladder cancer |
14 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC22A2 in bladder cancer | [ 7 ] | |||
|
Location |
TSS1500 (cg07026448) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.37E+00 | Statistic Test | p-value: 6.51E-06; Z-score: 1.21E+01 | ||
|
Methylation in Case |
6.18E-01 (Median) | Methylation in Control | 4.49E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC22A2 in bladder cancer | [ 7 ] | |||
|
Location |
TSS1500 (cg23995605) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.05E+00 | Statistic Test | p-value: 2.05E-04; Z-score: -3.50E+00 | ||
|
Methylation in Case |
8.82E-01 (Median) | Methylation in Control | 9.24E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC22A2 in bladder cancer | [ 7 ] | |||
|
Location |
TSS200 (cg19627213) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.57E+00 | Statistic Test | p-value: 3.81E-05; Z-score: -4.72E+00 | ||
|
Methylation in Case |
3.93E-01 (Median) | Methylation in Control | 6.18E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC22A2 in bladder cancer | [ 7 ] | |||
|
Location |
TSS200 (cg06236276) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.11E+00 | Statistic Test | p-value: 5.91E-03; Z-score: -1.99E+00 | ||
|
Methylation in Case |
5.95E-01 (Median) | Methylation in Control | 6.61E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC22A2 in bladder cancer | [ 7 ] | |||
|
Location |
TSS200 (cg04294894) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.06E+00 | Statistic Test | p-value: 1.30E-02; Z-score: -2.26E+00 | ||
|
Methylation in Case |
7.62E-01 (Median) | Methylation in Control | 8.06E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC22A2 in bladder cancer | [ 7 ] | |||
|
Location |
TSS200 (cg26939503) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.17E+00 | Statistic Test | p-value: 1.32E-02; Z-score: -4.78E+00 | ||
|
Methylation in Case |
5.51E-01 (Median) | Methylation in Control | 6.43E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC22A2 in bladder cancer | [ 7 ] | |||
|
Location |
TSS200 (cg02666489) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.08E+00 | Statistic Test | p-value: 1.97E-02; Z-score: -1.90E+00 | ||
|
Methylation in Case |
6.54E-01 (Median) | Methylation in Control | 7.06E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC22A2 in bladder cancer | [ 7 ] | |||
|
Location |
1stExon (cg27204616) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.06E+00 | Statistic Test | p-value: 2.44E-03; Z-score: -4.31E+00 | ||
|
Methylation in Case |
9.01E-01 (Median) | Methylation in Control | 9.52E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLC22A2 in bladder cancer | [ 7 ] | |||
|
Location |
1stExon (cg19561774) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.03E+00 | Statistic Test | p-value: 2.75E-03; Z-score: -1.71E+00 | ||
|
Methylation in Case |
8.25E-01 (Median) | Methylation in Control | 8.48E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 10 |
Methylation of SLC22A2 in bladder cancer | [ 7 ] | |||
|
Location |
1stExon (cg13717233) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.03E+00 | Statistic Test | p-value: 1.21E-02; Z-score: -1.31E+00 | ||
|
Methylation in Case |
8.24E-01 (Median) | Methylation in Control | 8.45E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 11 |
Methylation of SLC22A2 in bladder cancer | [ 7 ] | |||
|
Location |
Body (cg14918008) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.32E+00 | Statistic Test | p-value: 8.47E-10; Z-score: 8.08E+00 | ||
|
Methylation in Case |
7.69E-01 (Median) | Methylation in Control | 5.84E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 12 |
Methylation of SLC22A2 in bladder cancer | [ 7 ] | |||
|
Location |
Body (cg04310488) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.66E+00 | Statistic Test | p-value: 1.06E-05; Z-score: -3.87E+00 | ||
|
Methylation in Case |
3.84E-01 (Median) | Methylation in Control | 6.40E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 13 |
Methylation of SLC22A2 in bladder cancer | [ 7 ] | |||
|
Location |
Body (cg02490934) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.48E+00 | Statistic Test | p-value: 1.13E-05; Z-score: -4.29E+00 | ||
|
Methylation in Case |
4.75E-01 (Median) | Methylation in Control | 7.03E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 14 |
Methylation of SLC22A2 in bladder cancer | [ 7 ] | |||
|
Location |
Body (cg07601258) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.04E+00 | Statistic Test | p-value: 3.33E-03; Z-score: -3.43E+00 | ||
|
Methylation in Case |
8.79E-01 (Median) | Methylation in Control | 9.12E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Breast cancer |
19 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC22A2 in breast cancer | [ 8 ] | |||
|
Location |
TSS1500 (cg14450766) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.50E+00 | Statistic Test | p-value: 3.71E-15; Z-score: -1.93E+00 | ||
|
Methylation in Case |
2.08E-01 (Median) | Methylation in Control | 3.11E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC22A2 in breast cancer | [ 8 ] | |||
|
Location |
TSS1500 (cg23995605) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.13E+00 | Statistic Test | p-value: 3.50E-14; Z-score: -5.03E+00 | ||
|
Methylation in Case |
7.86E-01 (Median) | Methylation in Control | 8.87E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC22A2 in breast cancer | [ 8 ] | |||
|
Location |
TSS1500 (cg19499998) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.11E+00 | Statistic Test | p-value: 5.42E-13; Z-score: -3.65E+00 | ||
|
Methylation in Case |
8.06E-01 (Median) | Methylation in Control | 8.96E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC22A2 in breast cancer | [ 8 ] | |||
|
Location |
TSS1500 (cg25545503) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.09E+00 | Statistic Test | p-value: 1.14E-09; Z-score: -1.80E+00 | ||
|
Methylation in Case |
8.54E-01 (Median) | Methylation in Control | 9.32E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC22A2 in breast cancer | [ 8 ] | |||
|
Location |
TSS200 (cg26939503) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.54E+00 | Statistic Test | p-value: 3.15E-20; Z-score: -4.08E+00 | ||
|
Methylation in Case |
4.32E-01 (Median) | Methylation in Control | 6.67E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC22A2 in breast cancer | [ 8 ] | |||
|
Location |
TSS200 (cg06236276) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.32E+00 | Statistic Test | p-value: 1.62E-19; Z-score: -4.20E+00 | ||
|
Methylation in Case |
5.03E-01 (Median) | Methylation in Control | 6.66E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC22A2 in breast cancer | [ 8 ] | |||
|
Location |
TSS200 (cg04294894) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.40E+00 | Statistic Test | p-value: 1.45E-16; Z-score: -3.33E+00 | ||
|
Methylation in Case |
5.62E-01 (Median) | Methylation in Control | 7.89E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC22A2 in breast cancer | [ 8 ] | |||
|
Location |
TSS200 (cg19627213) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.51E+00 | Statistic Test | p-value: 8.30E-14; Z-score: -1.97E+00 | ||
|
Methylation in Case |
3.98E-01 (Median) | Methylation in Control | 6.01E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLC22A2 in breast cancer | [ 8 ] | |||
|
Location |
TSS200 (cg21755969) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.16E+00 | Statistic Test | p-value: 1.07E-08; Z-score: -2.33E+00 | ||
|
Methylation in Case |
4.74E-01 (Median) | Methylation in Control | 5.52E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 10 |
Methylation of SLC22A2 in breast cancer | [ 8 ] | |||
|
Location |
TSS200 (cg02666489) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.07E+00 | Statistic Test | p-value: 5.03E-08; Z-score: -1.06E+00 | ||
|
Methylation in Case |
6.99E-01 (Median) | Methylation in Control | 7.47E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 11 |
Methylation of SLC22A2 in breast cancer | [ 8 ] | |||
|
Location |
1stExon (cg13717233) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.11E+00 | Statistic Test | p-value: 5.12E-13; Z-score: -4.25E+00 | ||
|
Methylation in Case |
7.54E-01 (Median) | Methylation in Control | 8.36E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 12 |
Methylation of SLC22A2 in breast cancer | [ 8 ] | |||
|
Location |
1stExon (cg27204616) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.15E+00 | Statistic Test | p-value: 2.39E-12; Z-score: -6.29E+00 | ||
|
Methylation in Case |
8.15E-01 (Median) | Methylation in Control | 9.36E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 13 |
Methylation of SLC22A2 in breast cancer | [ 8 ] | |||
|
Location |
1stExon (cg19561774) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.09E+00 | Statistic Test | p-value: 1.15E-09; Z-score: -1.89E+00 | ||
|
Methylation in Case |
7.54E-01 (Median) | Methylation in Control | 8.18E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 14 |
Methylation of SLC22A2 in breast cancer | [ 8 ] | |||
|
Location |
Body (cg17098387) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.30E+00 | Statistic Test | p-value: 6.95E-20; Z-score: -3.61E+00 | ||
|
Methylation in Case |
5.59E-01 (Median) | Methylation in Control | 7.27E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 15 |
Methylation of SLC22A2 in breast cancer | [ 8 ] | |||
|
Location |
Body (cg04310488) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.34E+00 | Statistic Test | p-value: 5.15E-15; Z-score: -2.69E+00 | ||
|
Methylation in Case |
5.64E-01 (Median) | Methylation in Control | 7.58E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 16 |
Methylation of SLC22A2 in breast cancer | [ 8 ] | |||
|
Location |
Body (cg14918008) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.13E+00 | Statistic Test | p-value: 2.00E-11; Z-score: -1.81E+00 | ||
|
Methylation in Case |
6.82E-01 (Median) | Methylation in Control | 7.72E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 17 |
Methylation of SLC22A2 in breast cancer | [ 8 ] | |||
|
Location |
Body (cg03488613) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.09E+00 | Statistic Test | p-value: 8.15E-10; Z-score: -2.01E+00 | ||
|
Methylation in Case |
8.03E-01 (Median) | Methylation in Control | 8.75E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 18 |
Methylation of SLC22A2 in breast cancer | [ 8 ] | |||
|
Location |
Body (cg07601258) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.05E+00 | Statistic Test | p-value: 2.64E-09; Z-score: -1.64E+00 | ||
|
Methylation in Case |
8.43E-01 (Median) | Methylation in Control | 8.85E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 19 |
Methylation of SLC22A2 in breast cancer | [ 8 ] | |||
|
Location |
Body (cg02490934) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.21E+00 | Statistic Test | p-value: 8.82E-03; Z-score: -1.05E+00 | ||
|
Methylation in Case |
4.20E-01 (Median) | Methylation in Control | 5.09E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Renal cell carcinoma |
4 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC22A2 in clear cell renal cell carcinoma | [ 9 ] | |||
|
Location |
TSS1500 (cg23995605) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.00E+00 | Statistic Test | p-value: 3.94E-04; Z-score: -2.90E-02 | ||
|
Methylation in Case |
9.23E-01 (Median) | Methylation in Control | 9.24E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC22A2 in clear cell renal cell carcinoma | [ 9 ] | |||
|
Location |
TSS200 (cg04294894) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.10E+00 | Statistic Test | p-value: 3.59E-02; Z-score: -1.28E+00 | ||
|
Methylation in Case |
6.05E-01 (Median) | Methylation in Control | 6.65E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC22A2 in clear cell renal cell carcinoma | [ 9 ] | |||
|
Location |
Body (cg02490934) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.28E+00 | Statistic Test | p-value: 3.42E-03; Z-score: -2.82E+00 | ||
|
Methylation in Case |
4.94E-01 (Median) | Methylation in Control | 6.34E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC22A2 in clear cell renal cell carcinoma | [ 9 ] | |||
|
Location |
Body (cg07601258) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.06E+00 | Statistic Test | p-value: 6.36E-03; Z-score: -9.85E-01 | ||
|
Methylation in Case |
7.68E-01 (Median) | Methylation in Control | 8.14E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colon cancer |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC22A2 in colon adenocarcinoma | [ 10 ] | |||
|
Location |
TSS1500 (cg09595245) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.16E+00 | Statistic Test | p-value: 5.57E-04; Z-score: -2.37E+00 | ||
|
Methylation in Case |
6.72E-01 (Median) | Methylation in Control | 7.80E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC22A2 in colon adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg05672174) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.09E+00 | Statistic Test | p-value: 9.28E-05; Z-score: -2.18E+00 | ||
|
Methylation in Case |
6.99E-01 (Median) | Methylation in Control | 7.60E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC22A2 in colon adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg26907313) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.15E+00 | Statistic Test | p-value: 1.00E-03; Z-score: -3.03E+00 | ||
|
Methylation in Case |
7.19E-01 (Median) | Methylation in Control | 8.25E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Depression |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC22A2 in depression | [ 11 ] | |||
|
Location |
TSS1500 (cg07026448) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.02E+00 | Statistic Test | p-value: 4.21E-02; Z-score: 1.81E-01 | ||
|
Methylation in Case |
4.21E-01 (Median) | Methylation in Control | 4.14E-01 (Median) | ||
|
Studied Phenotype |
Depression [ ICD-11: 6A8Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
HIV infection |
8 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC22A2 in HIV infection | [ 13 ] | |||
|
Location |
TSS1500 (cg14450766) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.07E+00 | Statistic Test | p-value: 6.14E-04; Z-score: -3.43E-01 | ||
|
Methylation in Case |
2.51E-01 (Median) | Methylation in Control | 2.69E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC22A2 in HIV infection | [ 13 ] | |||
|
Location |
TSS1500 (cg19499998) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.01E+00 | Statistic Test | p-value: 7.98E-04; Z-score: 7.92E-01 | ||
|
Methylation in Case |
9.33E-01 (Median) | Methylation in Control | 9.20E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC22A2 in HIV infection | [ 13 ] | |||
|
Location |
TSS200 (cg19627213) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.15E+00 | Statistic Test | p-value: 4.26E-06; Z-score: -1.45E+00 | ||
|
Methylation in Case |
5.04E-01 (Median) | Methylation in Control | 5.80E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC22A2 in HIV infection | [ 13 ] | |||
|
Location |
TSS200 (cg04294894) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.03E+00 | Statistic Test | p-value: 3.10E-03; Z-score: -6.46E-01 | ||
|
Methylation in Case |
8.24E-01 (Median) | Methylation in Control | 8.48E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC22A2 in HIV infection | [ 13 ] | |||
|
Location |
TSS200 (cg06236276) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.03E+00 | Statistic Test | p-value: 3.82E-02; Z-score: -4.79E-01 | ||
|
Methylation in Case |
6.56E-01 (Median) | Methylation in Control | 6.77E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC22A2 in HIV infection | [ 13 ] | |||
|
Location |
1stExon (cg19561774) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.03E+00 | Statistic Test | p-value: 2.37E-06; Z-score: 1.25E+00 | ||
|
Methylation in Case |
8.94E-01 (Median) | Methylation in Control | 8.69E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC22A2 in HIV infection | [ 13 ] | |||
|
Location |
Body (cg17098387) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.08E+00 | Statistic Test | p-value: 2.08E-04; Z-score: -1.02E+00 | ||
|
Methylation in Case |
6.62E-01 (Median) | Methylation in Control | 7.17E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC22A2 in HIV infection | [ 13 ] | |||
|
Location |
Body (cg02490934) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.08E+00 | Statistic Test | p-value: 2.97E-04; Z-score: -1.08E+00 | ||
|
Methylation in Case |
7.76E-01 (Median) | Methylation in Control | 8.34E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Lung adenocarcinoma |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC22A2 in lung adenocarcinoma | [ 14 ] | |||
|
Location |
TSS1500 (cg07026448) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.06E+00 | Statistic Test | p-value: 3.92E-02; Z-score: 7.17E-01 | ||
|
Methylation in Case |
6.11E-01 (Median) | Methylation in Control | 5.78E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC22A2 in lung adenocarcinoma | [ 14 ] | |||
|
Location |
TSS200 (cg19627213) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.12E+00 | Statistic Test | p-value: 3.43E-02; Z-score: -3.14E+00 | ||
|
Methylation in Case |
5.50E-01 (Median) | Methylation in Control | 6.17E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC22A2 in lung adenocarcinoma | [ 14 ] | |||
|
Location |
Body (cg17098387) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.11E+00 | Statistic Test | p-value: 2.72E-02; Z-score: -3.43E+00 | ||
|
Methylation in Case |
6.86E-01 (Median) | Methylation in Control | 7.62E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Papillary thyroid cancer |
13 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC22A2 in papillary thyroid cancer | [ 15 ] | |||
|
Location |
TSS1500 (cg25545503) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.06E+00 | Statistic Test | p-value: 6.84E-11; Z-score: 1.47E+00 | ||
|
Methylation in Case |
8.75E-01 (Median) | Methylation in Control | 8.23E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC22A2 in papillary thyroid cancer | [ 15 ] | |||
|
Location |
TSS1500 (cg19499998) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.07E+00 | Statistic Test | p-value: 1.17E-10; Z-score: 1.56E+00 | ||
|
Methylation in Case |
8.57E-01 (Median) | Methylation in Control | 8.01E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC22A2 in papillary thyroid cancer | [ 15 ] | |||
|
Location |
TSS1500 (cg23995605) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.03E+00 | Statistic Test | p-value: 1.64E-09; Z-score: -2.12E+00 | ||
|
Methylation in Case |
9.27E-01 (Median) | Methylation in Control | 9.53E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC22A2 in papillary thyroid cancer | [ 15 ] | |||
|
Location |
TSS1500 (cg07026448) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.08E+00 | Statistic Test | p-value: 9.62E-03; Z-score: 1.09E+00 | ||
|
Methylation in Case |
6.88E-01 (Median) | Methylation in Control | 6.37E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC22A2 in papillary thyroid cancer | [ 15 ] | |||
|
Location |
TSS200 (cg06236276) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.06E+00 | Statistic Test | p-value: 1.58E-06; Z-score: -1.16E+00 | ||
|
Methylation in Case |
6.87E-01 (Median) | Methylation in Control | 7.30E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC22A2 in papillary thyroid cancer | [ 15 ] | |||
|
Location |
TSS200 (cg26939503) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.05E+00 | Statistic Test | p-value: 2.36E-05; Z-score: -6.23E-01 | ||
|
Methylation in Case |
6.43E-01 (Median) | Methylation in Control | 6.72E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC22A2 in papillary thyroid cancer | [ 15 ] | |||
|
Location |
TSS200 (cg02666489) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.10E+00 | Statistic Test | p-value: 5.04E-05; Z-score: 1.73E+00 | ||
|
Methylation in Case |
8.37E-01 (Median) | Methylation in Control | 7.62E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC22A2 in papillary thyroid cancer | [ 15 ] | |||
|
Location |
TSS200 (cg19627213) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.06E+00 | Statistic Test | p-value: 3.42E-03; Z-score: -5.49E-01 | ||
|
Methylation in Case |
6.66E-01 (Median) | Methylation in Control | 7.03E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLC22A2 in papillary thyroid cancer | [ 15 ] | |||
|
Location |
1stExon (cg19561774) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.02E+00 | Statistic Test | p-value: 5.02E-09; Z-score: -1.47E+00 | ||
|
Methylation in Case |
9.20E-01 (Median) | Methylation in Control | 9.37E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 10 |
Methylation of SLC22A2 in papillary thyroid cancer | [ 15 ] | |||
|
Location |
1stExon (cg27204616) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.02E+00 | Statistic Test | p-value: 1.60E-06; Z-score: -1.22E+00 | ||
|
Methylation in Case |
9.01E-01 (Median) | Methylation in Control | 9.18E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 11 |
Methylation of SLC22A2 in papillary thyroid cancer | [ 15 ] | |||
|
Location |
1stExon (cg13717233) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.02E+00 | Statistic Test | p-value: 3.30E-06; Z-score: -9.37E-01 | ||
|
Methylation in Case |
8.97E-01 (Median) | Methylation in Control | 9.11E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 12 |
Methylation of SLC22A2 in papillary thyroid cancer | [ 15 ] | |||
|
Location |
Body (cg02490934) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.07E+00 | Statistic Test | p-value: 7.43E-06; Z-score: -1.20E+00 | ||
|
Methylation in Case |
7.82E-01 (Median) | Methylation in Control | 8.36E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 13 |
Methylation of SLC22A2 in papillary thyroid cancer | [ 15 ] | |||
|
Location |
Body (cg07601258) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.00E+00 | Statistic Test | p-value: 8.06E-04; Z-score: -2.45E-01 | ||
|
Methylation in Case |
9.40E-01 (Median) | Methylation in Control | 9.43E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Systemic lupus erythematosus |
7 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC22A2 in systemic lupus erythematosus | [ 16 ] | |||
|
Location |
TSS1500 (cg23995605) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.00E+00 | Statistic Test | p-value: 2.95E-02; Z-score: -2.21E-01 | ||
|
Methylation in Case |
9.19E-01 (Median) | Methylation in Control | 9.24E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus [ ICD-11: 4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC22A2 in systemic lupus erythematosus | [ 16 ] | |||
|
Location |
TSS200 (cg26939503) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.03E+00 | Statistic Test | p-value: 1.19E-03; Z-score: -2.66E-01 | ||
|
Methylation in Case |
6.52E-01 (Median) | Methylation in Control | 6.72E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus [ ICD-11: 4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC22A2 in systemic lupus erythematosus | [ 16 ] | |||
|
Location |
TSS200 (cg02666489) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.01E+00 | Statistic Test | p-value: 1.35E-02; Z-score: -2.51E-01 | ||
|
Methylation in Case |
8.70E-01 (Median) | Methylation in Control | 8.77E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus [ ICD-11: 4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC22A2 in systemic lupus erythematosus | [ 16 ] | |||
|
Location |
TSS200 (cg06236276) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.01E+00 | Statistic Test | p-value: 3.12E-02; Z-score: -1.25E-01 | ||
|
Methylation in Case |
7.03E-01 (Median) | Methylation in Control | 7.11E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus [ ICD-11: 4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC22A2 in systemic lupus erythematosus | [ 16 ] | |||
|
Location |
Body (cg07601258) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.00E+00 | Statistic Test | p-value: 5.79E-03; Z-score: -2.06E-01 | ||
|
Methylation in Case |
9.24E-01 (Median) | Methylation in Control | 9.28E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus [ ICD-11: 4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC22A2 in systemic lupus erythematosus | [ 16 ] | |||
|
Location |
Body (cg17098387) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.02E+00 | Statistic Test | p-value: 7.48E-03; Z-score: -1.85E-01 | ||
|
Methylation in Case |
6.55E-01 (Median) | Methylation in Control | 6.69E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus [ ICD-11: 4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC22A2 in systemic lupus erythematosus | [ 16 ] | |||
|
Location |
Body (cg02490934) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.02E+00 | Statistic Test | p-value: 3.70E-02; Z-score: -1.77E-01 | ||
|
Methylation in Case |
7.69E-01 (Median) | Methylation in Control | 7.85E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus [ ICD-11: 4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Panic disorder |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC22A2 in panic disorder | [ 17 ] | |||
|
Location |
TSS200 (cg04294894) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.08E+00 | Statistic Test | p-value: 3.35E-02; Z-score: 4.99E-01 | ||
|
Methylation in Case |
2.72E+00 (Median) | Methylation in Control | 2.53E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder [ ICD-11: 6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC22A2 in panic disorder | [ 17 ] | |||
|
Location |
TSS200 (cg21755969) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.30E+00 | Statistic Test | p-value: 3.64E-02; Z-score: 4.24E-01 | ||
|
Methylation in Case |
6.92E-01 (Median) | Methylation in Control | 5.31E-01 (Median) | ||
|
Studied Phenotype |
Panic disorder [ ICD-11: 6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Atypical teratoid rhabdoid tumor |
7 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC22A2 in atypical teratoid rhabdoid tumor | [ 18 ] | |||
|
Location |
1stExon (cg13717233) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.49E+00 | Statistic Test | p-value: 2.58E-19; Z-score: -3.07E+00 | ||
|
Methylation in Case |
5.11E-01 (Median) | Methylation in Control | 7.62E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC22A2 in atypical teratoid rhabdoid tumor | [ 18 ] | |||
|
Location |
1stExon (cg19561774) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.18E+00 | Statistic Test | p-value: 1.13E-17; Z-score: -2.79E+00 | ||
|
Methylation in Case |
7.34E-01 (Median) | Methylation in Control | 8.65E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC22A2 in atypical teratoid rhabdoid tumor | [ 18 ] | |||
|
Location |
1stExon (cg27204616) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.36E+00 | Statistic Test | p-value: 9.27E-16; Z-score: -3.43E+00 | ||
|
Methylation in Case |
5.38E-01 (Median) | Methylation in Control | 7.33E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC22A2 in atypical teratoid rhabdoid tumor | [ 18 ] | |||
|
Location |
Body (cg02490934) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.03E+00 | Statistic Test | p-value: 1.67E-04; Z-score: -7.24E-01 | ||
|
Methylation in Case |
8.91E-01 (Median) | Methylation in Control | 9.16E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC22A2 in atypical teratoid rhabdoid tumor | [ 18 ] | |||
|
Location |
Body (cg03488613) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.33E+00 | Statistic Test | p-value: 3.52E-04; Z-score: -1.02E+00 | ||
|
Methylation in Case |
5.67E-01 (Median) | Methylation in Control | 7.52E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC22A2 in atypical teratoid rhabdoid tumor | [ 18 ] | |||
|
Location |
Body (cg04310488) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.04E+00 | Statistic Test | p-value: 7.00E-04; Z-score: -4.47E-01 | ||
|
Methylation in Case |
7.84E-01 (Median) | Methylation in Control | 8.16E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC22A2 in atypical teratoid rhabdoid tumor | [ 18 ] | |||
|
Location |
Body (cg07601258) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.06E+00 | Statistic Test | p-value: 4.49E-03; Z-score: 6.32E-01 | ||
|
Methylation in Case |
9.30E-01 (Median) | Methylation in Control | 8.75E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colorectal cancer |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC22A2 in colorectal cancer | [ 19 ] | |||
|
Location |
1stExon (cg19561774) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.00E+00 | Statistic Test | p-value: 8.06E-03; Z-score: -1.88E-01 | ||
|
Methylation in Case |
9.48E-01 (Median) | Methylation in Control | 9.50E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC22A2 in colorectal cancer | [ 19 ] | |||
|
Location |
Body (cg03488613) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.03E+00 | Statistic Test | p-value: 9.96E-03; Z-score: 9.12E-01 | ||
|
Methylation in Case |
9.20E-01 (Median) | Methylation in Control | 8.94E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC22A2 in colorectal cancer | [ 19 ] | |||
|
Location |
Body (cg07601258) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.01E+00 | Statistic Test | p-value: 2.47E-02; Z-score: 3.07E-01 | ||
|
Methylation in Case |
9.45E-01 (Median) | Methylation in Control | 9.40E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Pancretic ductal adenocarcinoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC22A2 in pancretic ductal adenocarcinoma | [ 20 ] | |||
|
Location |
Body (cg12933431) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.26E+00 | Statistic Test | p-value: 1.75E-03; Z-score: 1.32E+00 | ||
|
Methylation in Case |
6.71E-01 (Median) | Methylation in Control | 5.32E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Esthesioneuroblastoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Moderate hypomethylation of SLC22A2 in esthesioneuroblastoma than that in healthy individual | ||||
Studied Phenotype |
Esthesioneuroblastoma [ICD-11:2D50.1] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 1.63E-05; Fold-change: -0.213965366; Z-score: -1.711614709 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
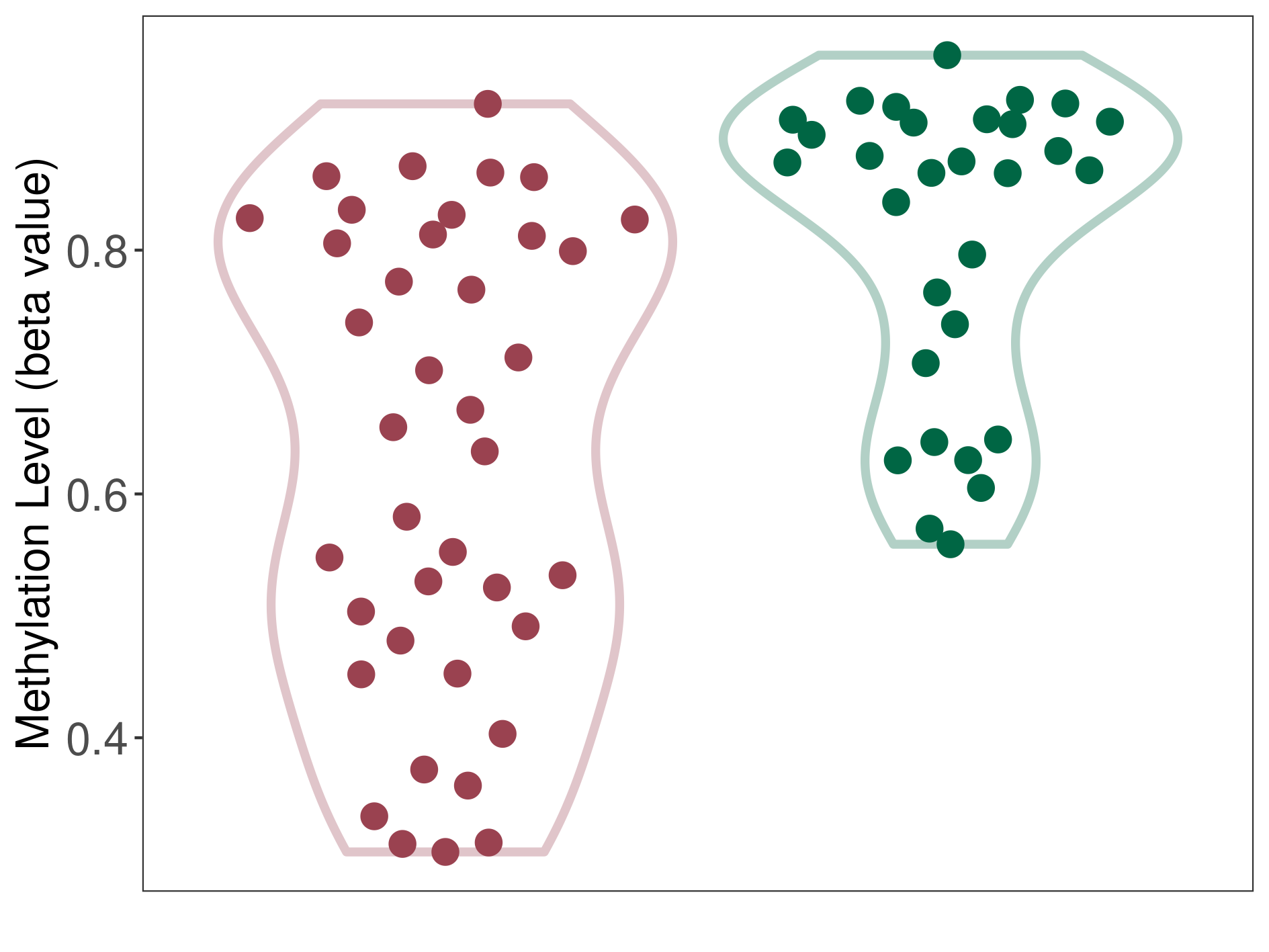
|
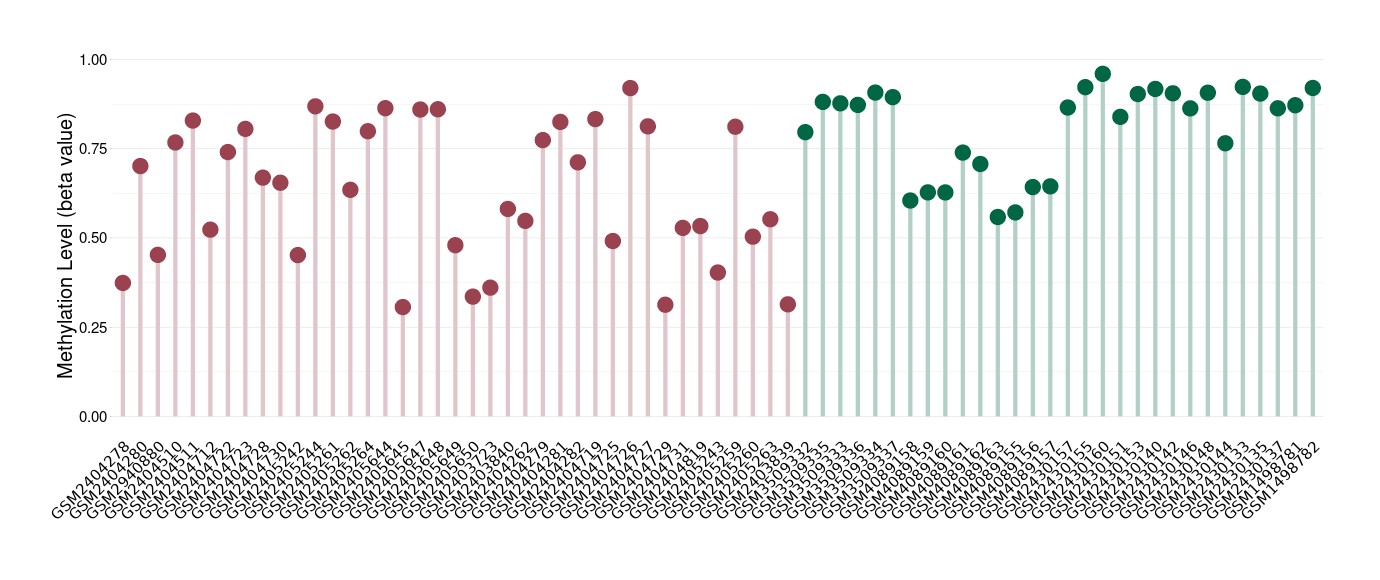 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Medulloblastoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Moderate hypomethylation of SLC22A2 in medulloblastoma than that in healthy individual | ||||
Studied Phenotype |
Medulloblastoma [ICD-11:2A00.10] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 6.98E-86; Fold-change: -0.263759902; Z-score: -3.445624036 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Melanoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Moderate hypomethylation of SLC22A2 in melanoma than that in healthy individual | ||||
Studied Phenotype |
Melanoma [ICD-11:2C30] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 0.002420386; Fold-change: -0.241840851; Z-score: -1.569377238 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Multilayered rosettes embryonal tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Moderate hypomethylation of SLC22A2 in multilayered rosettes embryonal tumour than that in healthy individual | ||||
Studied Phenotype |
Multilayered rosettes embryonal tumour [ICD-11:2A00.1] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 1.37E-09; Fold-change: -0.261324171; Z-score: -2.25939206 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||

|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Cerebellar liponeurocytoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypomethylation of SLC22A2 in cerebellar liponeurocytoma than that in healthy individual | ||||
Studied Phenotype |
Cerebellar liponeurocytoma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 1.59E-08; Fold-change: -0.617671121; Z-score: -2.933433032 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
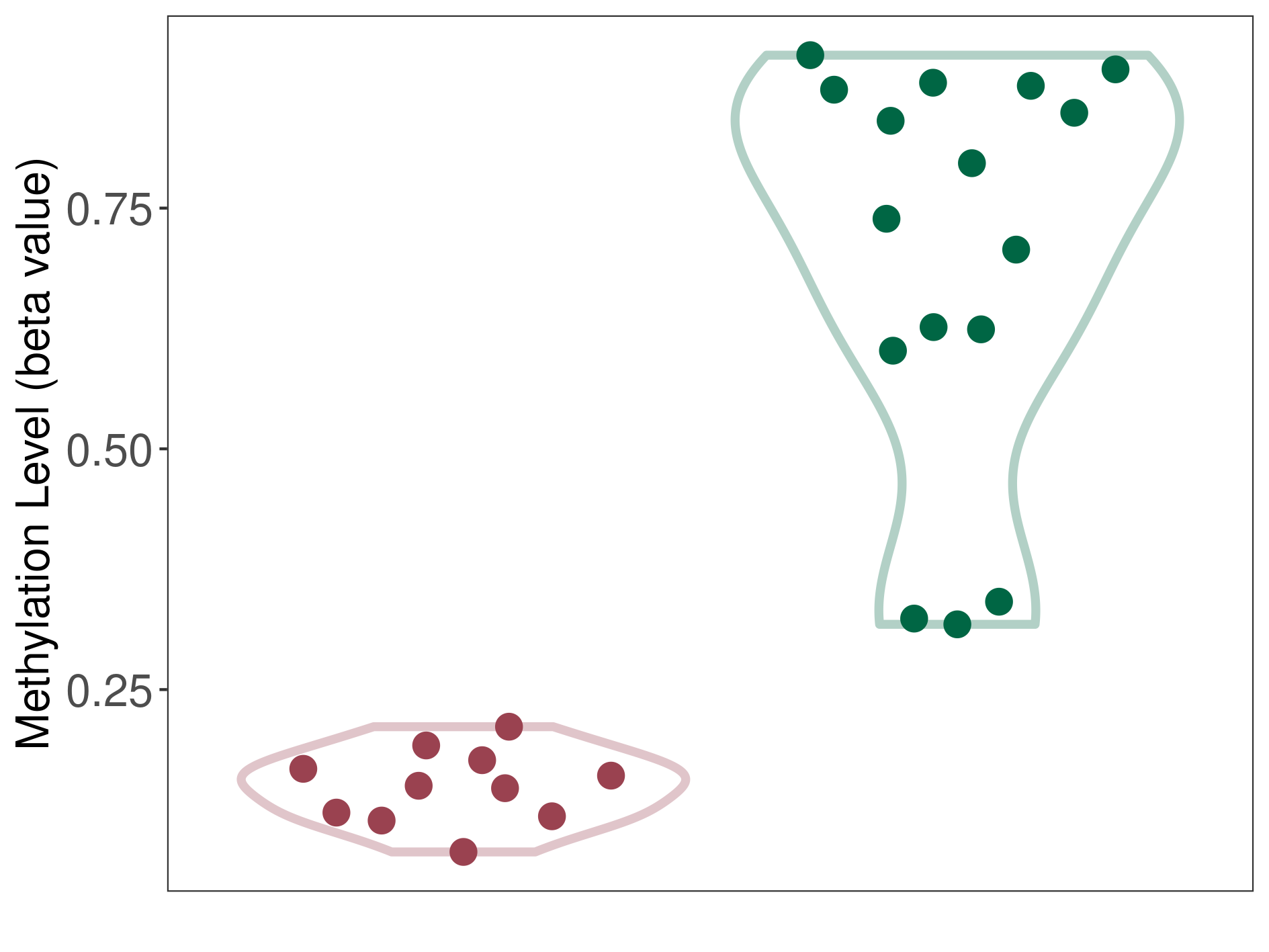
|
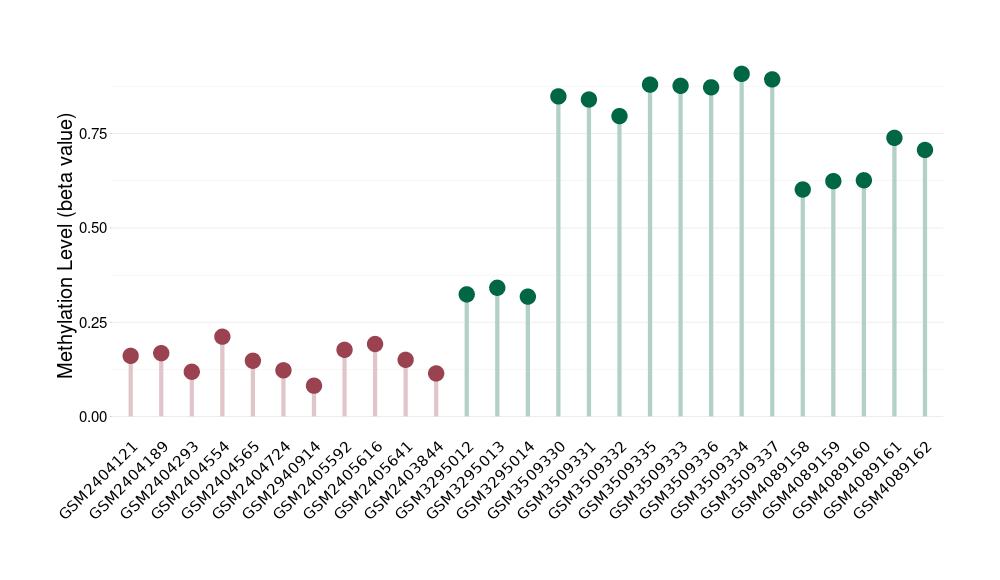 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Lymphoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypomethylation of SLC22A2 in lymphoma than that in healthy individual | ||||
Studied Phenotype |
Lymphoma [ICD-11:2B30] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 1.35E-08; Fold-change: -0.308339461; Z-score: -2.124187076 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
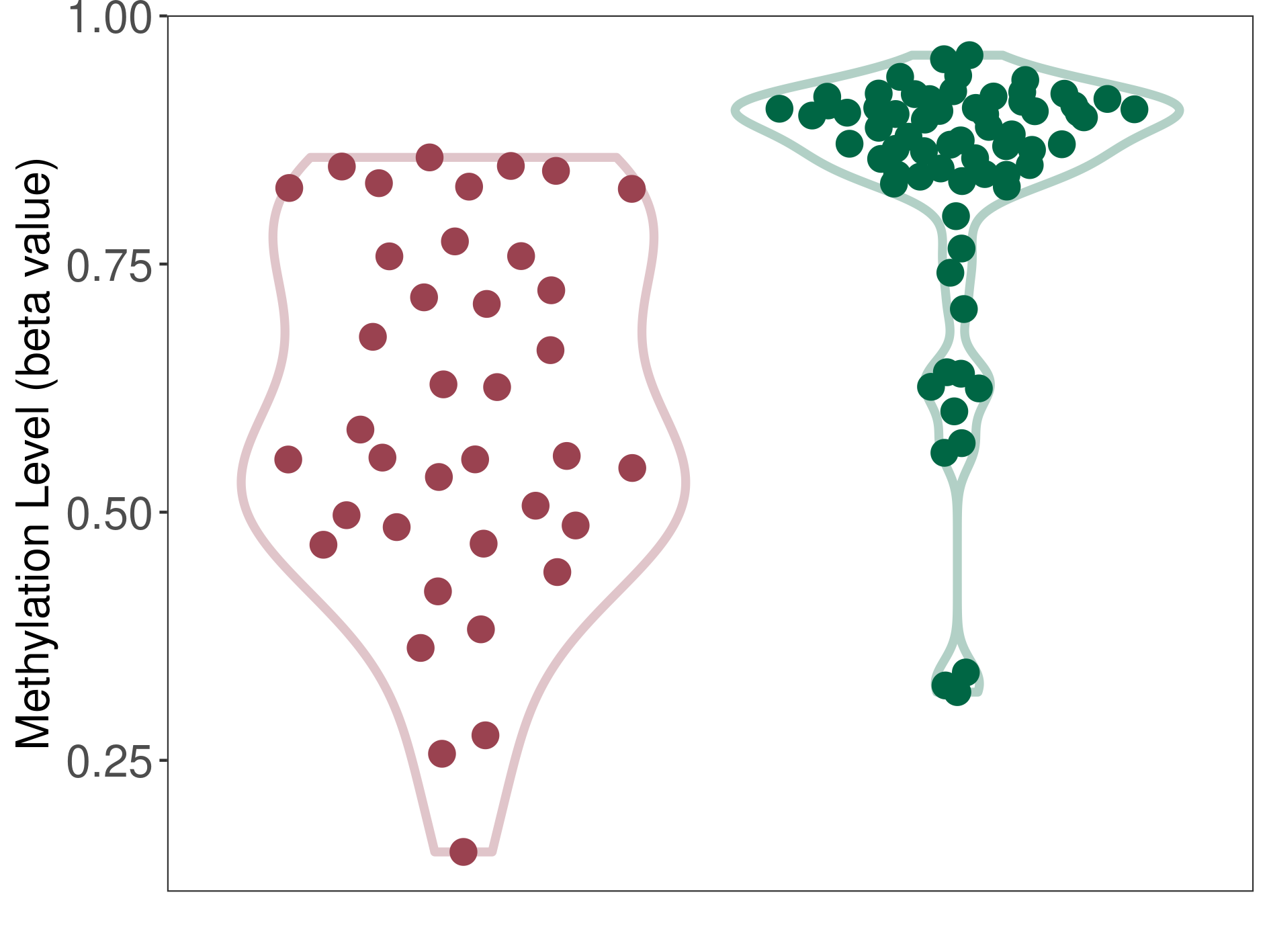
|
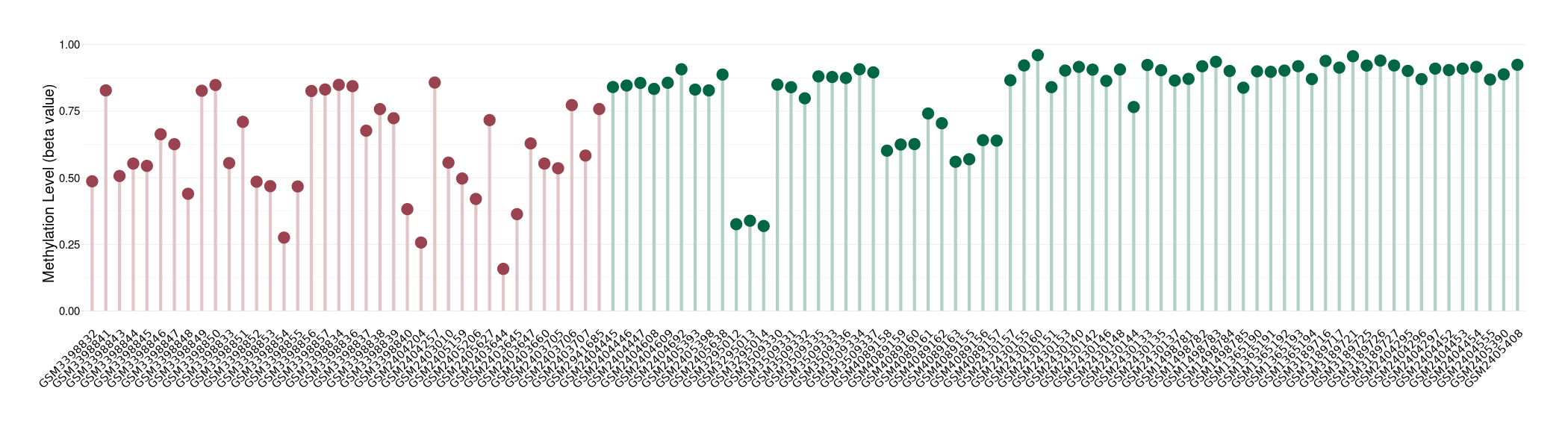 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Meningioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypomethylation of SLC22A2 in meningioma than that in healthy individual | ||||
Studied Phenotype |
Meningioma [ICD-11:2A01.0Z] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 1.25E-62; Fold-change: -0.340860889; Z-score: -3.217549796 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
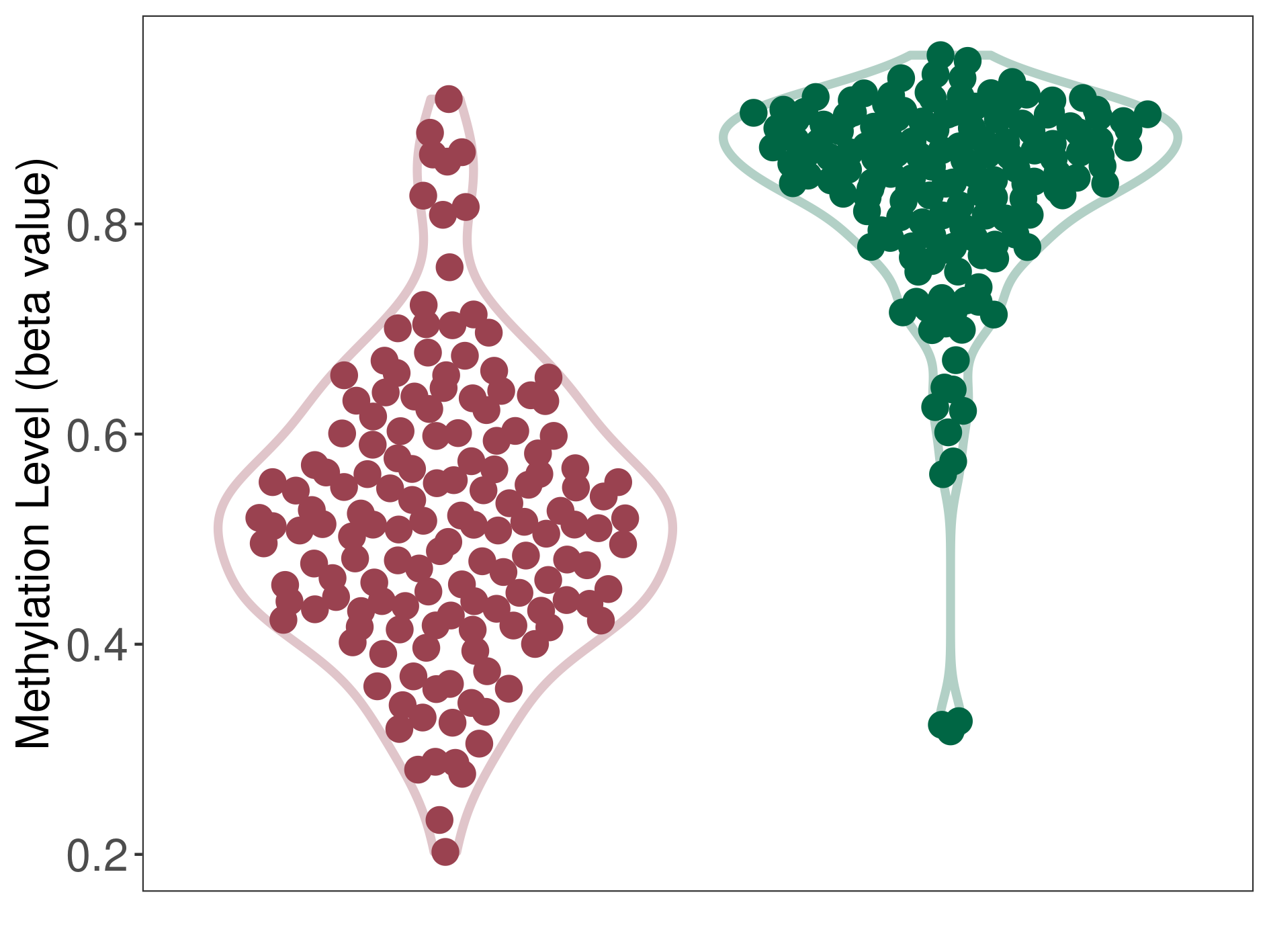
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Posterior fossa ependymoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypomethylation of SLC22A2 in posterior fossa ependymoma than that in healthy individual | ||||
Studied Phenotype |
Posterior fossa ependymoma [ICD-11:2D50.2] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 3.20E-102; Fold-change: -0.383390542; Z-score: -3.474081509 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||

|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Histone acetylation |
|||||
|
Renal cell carcinoma |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Hypoacetylation of SLC22A2 in renal cell carcinoma | [ 21 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes |
Down regulation of SLC22A2 | Experiment Method | RT-qPCR | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patients samples of human; Multiple cell lines of human | ||||
|
Additional Notes |
Combined treatment using the DNA methylation inhibitor decitabine and the histone deacetylase inhibitor vorinostat significantly increased the expression of OCT2 in RCC cell lines, which sensitized these cells to oxaliplatin. | ||||
|
Epigenetic Phenomenon 2 |
Hypoacetylation of SLC22A2 in renal cell carcinoma | [ 21 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes |
Down regulation of SLC22A2 | Experiment Method | RT-qPCR | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patients samples of human; Multiple cell lines of human | ||||
|
Additional Notes |
Combined treatment using the DNA methylation inhibitor decitabine and the histone deacetylase inhibitor vorinostat significantly increased the expression of OCT2 in RCC cell lines, which sensitized these cells to oxaliplatin. | ||||
|
microRNA |
|||||
|
Renal cell carcinoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
miR-21 silencing increased SLC22A2 expression | [ 22 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | . | ||
|
Related Molecular Changes |
Down regulation of SLC22A2 | Experiment Method | RT-qPCR | ||
|
miRNA Stemloop ID |
miR-21 | miRNA Mature ID | miR-21-5p | ||
|
miRNA Sequence |
UAGCUUAUCAGACUGAUGUUGA | ||||
|
miRNA Target Type |
Unknown | ||||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
Targeting miR-21 decreases expression of SLC22A2 and promotes chemosensitivity of renal carcinoma. | ||||
|
Unclear Phenotype |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
miR-335 directly targets SLC22A2 | [ 23 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Microarray | ||
|
miRNA Stemloop ID |
miR-335 | miRNA Mature ID | miR-335-5p | ||
|
miRNA Sequence |
UCAAGAGCAAUAACGAAAAAUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.
