Detail Information of Epigenetic Regulations
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0179 Transporter Info | ||||
| Gene Name | SLC25A2 | ||||
| Transporter Name | Mitochondrial ornithine transporter 2 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| Epigenetic Regulations of This DT (EGR) | |||||
|---|---|---|---|---|---|
|
Methylation |
|||||
|
Colon cancer |
11 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC25A2 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
5'UTR (cg04349243) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 2.31E+00 | Statistic Test | p-value: 7.69E-10; Z-score: 6.60E+00 | ||
|
Methylation in Case |
6.81E-01 (Median) | Methylation in Control | 2.95E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC25A2 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg04422990) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.49E+00 | Statistic Test | p-value: 2.31E-04; Z-score: 4.04E+00 | ||
|
Methylation in Case |
3.67E-01 (Median) | Methylation in Control | 2.47E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC25A2 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg04431946) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 2.16E+00 | Statistic Test | p-value: 2.69E-12; Z-score: 4.63E+00 | ||
|
Methylation in Case |
7.02E-01 (Median) | Methylation in Control | 3.25E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC25A2 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg04442576) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 2.80E+00 | Statistic Test | p-value: 2.26E-09; Z-score: 8.55E+00 | ||
|
Methylation in Case |
5.91E-01 (Median) | Methylation in Control | 2.11E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC25A2 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg12005098) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 3.69E+00 | Statistic Test | p-value: 1.54E-07; Z-score: 6.14E+00 | ||
|
Methylation in Case |
5.42E-01 (Median) | Methylation in Control | 1.47E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC25A2 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg22149516) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 2.47E+00 | Statistic Test | p-value: 7.10E-04; Z-score: 5.76E+00 | ||
|
Methylation in Case |
3.44E-01 (Median) | Methylation in Control | 1.39E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC25A2 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
1stExon (cg26708235) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 2.55E+00 | Statistic Test | p-value: 2.23E-08; Z-score: 4.70E+00 | ||
|
Methylation in Case |
5.68E-01 (Median) | Methylation in Control | 2.23E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC25A2 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg21324456) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -2.78E+00 | Statistic Test | p-value: 1.62E-12; Z-score: -4.88E+00 | ||
|
Methylation in Case |
1.23E-01 (Median) | Methylation in Control | 3.43E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLC25A2 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg01995743) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 2.73E+00 | Statistic Test | p-value: 5.05E-12; Z-score: 4.23E+00 | ||
|
Methylation in Case |
6.06E-01 (Median) | Methylation in Control | 2.22E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 10 |
Methylation of SLC25A2 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg20912978) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.74E+00 | Statistic Test | p-value: 6.08E-10; Z-score: -2.56E+00 | ||
|
Methylation in Case |
1.59E-01 (Median) | Methylation in Control | 2.76E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 11 |
Methylation of SLC25A2 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg03519481) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.10E+00 | Statistic Test | p-value: 7.83E-04; Z-score: -3.27E+00 | ||
|
Methylation in Case |
7.46E-01 (Median) | Methylation in Control | 8.23E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Bladder cancer |
12 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC25A2 in bladder cancer | [ 2 ] | |||
|
Location |
TSS1500 (cg26518745) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.04E+00 | Statistic Test | p-value: 9.29E-04; Z-score: -3.63E+00 | ||
|
Methylation in Case |
8.55E-01 (Median) | Methylation in Control | 8.92E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC25A2 in bladder cancer | [ 2 ] | |||
|
Location |
TSS1500 (cg26783353) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.03E+00 | Statistic Test | p-value: 2.64E-02; Z-score: 1.52E+00 | ||
|
Methylation in Case |
9.02E-01 (Median) | Methylation in Control | 8.76E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC25A2 in bladder cancer | [ 2 ] | |||
|
Location |
TSS200 (cg25212131) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.50E+00 | Statistic Test | p-value: 7.30E-11; Z-score: 9.76E+00 | ||
|
Methylation in Case |
8.44E-01 (Median) | Methylation in Control | 5.63E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC25A2 in bladder cancer | [ 2 ] | |||
|
Location |
TSS200 (cg00030420) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.25E+00 | Statistic Test | p-value: 1.07E-08; Z-score: 8.94E+00 | ||
|
Methylation in Case |
8.45E-01 (Median) | Methylation in Control | 6.78E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC25A2 in bladder cancer | [ 2 ] | |||
|
Location |
TSS200 (cg05845376) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 2.08E+00 | Statistic Test | p-value: 2.06E-08; Z-score: 1.05E+01 | ||
|
Methylation in Case |
9.08E-01 (Median) | Methylation in Control | 4.36E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC25A2 in bladder cancer | [ 2 ] | |||
|
Location |
TSS200 (cg27320005) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.42E+00 | Statistic Test | p-value: 1.28E-07; Z-score: 7.03E+00 | ||
|
Methylation in Case |
8.94E-01 (Median) | Methylation in Control | 6.32E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC25A2 in bladder cancer | [ 2 ] | |||
|
Location |
TSS200 (cg07039560) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.74E+00 | Statistic Test | p-value: 1.52E-06; Z-score: 6.13E+00 | ||
|
Methylation in Case |
7.63E-01 (Median) | Methylation in Control | 4.40E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC25A2 in bladder cancer | [ 2 ] | |||
|
Location |
TSS200 (cg07496545) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.76E+00 | Statistic Test | p-value: 4.63E-05; Z-score: 5.65E+00 | ||
|
Methylation in Case |
9.16E-01 (Median) | Methylation in Control | 5.22E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLC25A2 in bladder cancer | [ 2 ] | |||
|
Location |
TSS200 (cg21636683) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.47E+00 | Statistic Test | p-value: 7.77E-05; Z-score: 4.18E+00 | ||
|
Methylation in Case |
7.41E-01 (Median) | Methylation in Control | 5.05E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 10 |
Methylation of SLC25A2 in bladder cancer | [ 2 ] | |||
|
Location |
1stExon (cg05638493) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 2.22E+00 | Statistic Test | p-value: 1.19E-13; Z-score: 1.70E+01 | ||
|
Methylation in Case |
6.72E-01 (Median) | Methylation in Control | 3.03E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 11 |
Methylation of SLC25A2 in bladder cancer | [ 2 ] | |||
|
Location |
1stExon (cg03060468) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.33E+00 | Statistic Test | p-value: 8.42E-07; Z-score: 2.07E+01 | ||
|
Methylation in Case |
6.64E-01 (Median) | Methylation in Control | 5.00E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 12 |
Methylation of SLC25A2 in bladder cancer | [ 2 ] | |||
|
Location |
1stExon (cg02553537) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.02E+00 | Statistic Test | p-value: 2.03E-02; Z-score: -7.49E-01 | ||
|
Methylation in Case |
7.73E-01 (Median) | Methylation in Control | 7.90E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Renal cell carcinoma |
11 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC25A2 in clear cell renal cell carcinoma | [ 3 ] | |||
|
Location |
TSS1500 (cg22748470) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.06E+00 | Statistic Test | p-value: 1.24E-03; Z-score: 1.05E+00 | ||
|
Methylation in Case |
8.93E-01 (Median) | Methylation in Control | 8.46E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC25A2 in clear cell renal cell carcinoma | [ 3 ] | |||
|
Location |
TSS1500 (cg26518745) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.00E+00 | Statistic Test | p-value: 4.52E-03; Z-score: -4.95E-01 | ||
|
Methylation in Case |
9.56E-01 (Median) | Methylation in Control | 9.59E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC25A2 in clear cell renal cell carcinoma | [ 3 ] | |||
|
Location |
TSS200 (cg25212131) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.33E+00 | Statistic Test | p-value: 4.05E-21; Z-score: 3.84E+00 | ||
|
Methylation in Case |
7.79E-01 (Median) | Methylation in Control | 5.88E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC25A2 in clear cell renal cell carcinoma | [ 3 ] | |||
|
Location |
TSS200 (cg05845376) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.73E+00 | Statistic Test | p-value: 2.79E-18; Z-score: 2.89E+00 | ||
|
Methylation in Case |
8.73E-01 (Median) | Methylation in Control | 5.03E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC25A2 in clear cell renal cell carcinoma | [ 3 ] | |||
|
Location |
TSS200 (cg07496545) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.73E+00 | Statistic Test | p-value: 1.27E-17; Z-score: 2.87E+00 | ||
|
Methylation in Case |
8.73E-01 (Median) | Methylation in Control | 5.05E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC25A2 in clear cell renal cell carcinoma | [ 3 ] | |||
|
Location |
TSS200 (cg07039560) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.47E+00 | Statistic Test | p-value: 1.35E-15; Z-score: 3.37E+00 | ||
|
Methylation in Case |
6.40E-01 (Median) | Methylation in Control | 4.36E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC25A2 in clear cell renal cell carcinoma | [ 3 ] | |||
|
Location |
TSS200 (cg27320005) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.33E+00 | Statistic Test | p-value: 3.40E-15; Z-score: 3.25E+00 | ||
|
Methylation in Case |
8.49E-01 (Median) | Methylation in Control | 6.39E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC25A2 in clear cell renal cell carcinoma | [ 3 ] | |||
|
Location |
TSS200 (cg00030420) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.17E+00 | Statistic Test | p-value: 7.35E-14; Z-score: 3.49E+00 | ||
|
Methylation in Case |
9.20E-01 (Median) | Methylation in Control | 7.89E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLC25A2 in clear cell renal cell carcinoma | [ 3 ] | |||
|
Location |
TSS200 (cg21636683) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.28E+00 | Statistic Test | p-value: 1.54E-10; Z-score: 3.09E+00 | ||
|
Methylation in Case |
7.91E-01 (Median) | Methylation in Control | 6.18E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 10 |
Methylation of SLC25A2 in clear cell renal cell carcinoma | [ 3 ] | |||
|
Location |
1stExon (cg05638493) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.54E+00 | Statistic Test | p-value: 7.87E-09; Z-score: 3.89E+00 | ||
|
Methylation in Case |
5.35E-01 (Median) | Methylation in Control | 3.47E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 11 |
Methylation of SLC25A2 in clear cell renal cell carcinoma | [ 3 ] | |||
|
Location |
1stExon (cg03060468) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.11E+00 | Statistic Test | p-value: 4.98E-03; Z-score: 1.45E+00 | ||
|
Methylation in Case |
6.66E-01 (Median) | Methylation in Control | 6.02E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colorectal cancer |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC25A2 in colorectal cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg22748470) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.02E+00 | Statistic Test | p-value: 3.57E-02; Z-score: -5.25E-01 | ||
|
Methylation in Case |
8.33E-01 (Median) | Methylation in Control | 8.50E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC25A2 in colorectal cancer | [ 4 ] | |||
|
Location |
1stExon (cg02553537) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.02E+00 | Statistic Test | p-value: 1.60E-02; Z-score: -2.01E+00 | ||
|
Methylation in Case |
9.33E-01 (Median) | Methylation in Control | 9.49E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Hepatocellular carcinoma |
13 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC25A2 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg20361238) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.40E+00 | Statistic Test | p-value: 1.23E-15; Z-score: -4.10E+00 | ||
|
Methylation in Case |
5.55E-01 (Median) | Methylation in Control | 7.76E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC25A2 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg23623667) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 2.84E+00 | Statistic Test | p-value: 2.86E-15; Z-score: 6.88E+00 | ||
|
Methylation in Case |
3.66E-01 (Median) | Methylation in Control | 1.29E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC25A2 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg26518745) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.10E+00 | Statistic Test | p-value: 1.32E-07; Z-score: -1.74E+00 | ||
|
Methylation in Case |
7.60E-01 (Median) | Methylation in Control | 8.33E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC25A2 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg19215261) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.01E+00 | Statistic Test | p-value: 4.28E-03; Z-score: -5.82E-01 | ||
|
Methylation in Case |
8.87E-01 (Median) | Methylation in Control | 8.96E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC25A2 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg22748470) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.01E+00 | Statistic Test | p-value: 1.73E-02; Z-score: 1.40E-01 | ||
|
Methylation in Case |
8.23E-01 (Median) | Methylation in Control | 8.16E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC25A2 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS200 (cg27320005) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.22E+00 | Statistic Test | p-value: 9.55E-08; Z-score: 1.96E+00 | ||
|
Methylation in Case |
8.50E-01 (Median) | Methylation in Control | 6.98E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC25A2 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS200 (cg07496545) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.25E+00 | Statistic Test | p-value: 1.24E-05; Z-score: 1.72E+00 | ||
|
Methylation in Case |
8.44E-01 (Median) | Methylation in Control | 6.73E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC25A2 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
1stExon (cg02553537) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.05E+00 | Statistic Test | p-value: 1.64E-05; Z-score: -1.57E+00 | ||
|
Methylation in Case |
8.33E-01 (Median) | Methylation in Control | 8.72E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLC25A2 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
1stExon (cg03060468) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.13E+00 | Statistic Test | p-value: 2.68E-02; Z-score: 1.38E+00 | ||
|
Methylation in Case |
6.10E-01 (Median) | Methylation in Control | 5.41E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 10 |
Methylation of SLC25A2 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
Body (cg12377084) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.50E+00 | Statistic Test | p-value: 8.61E-19; Z-score: -8.13E+00 | ||
|
Methylation in Case |
5.38E-01 (Median) | Methylation in Control | 8.07E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 11 |
Methylation of SLC25A2 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
Body (cg13154880) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 2.06E+00 | Statistic Test | p-value: 1.14E-17; Z-score: 8.30E+00 | ||
|
Methylation in Case |
3.96E-01 (Median) | Methylation in Control | 1.92E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 12 |
Methylation of SLC25A2 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
Body (cg02675946) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -3.13E+00 | Statistic Test | p-value: 4.24E-14; Z-score: -2.09E+00 | ||
|
Methylation in Case |
9.90E-02 (Median) | Methylation in Control | 3.10E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 13 |
Methylation of SLC25A2 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
3'UTR (cg15401862) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.61E+00 | Statistic Test | p-value: 1.33E-18; Z-score: -4.49E+00 | ||
|
Methylation in Case |
4.47E-01 (Median) | Methylation in Control | 7.21E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Panic disorder |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC25A2 in panic disorder | [ 6 ] | |||
|
Location |
TSS1500 (cg26518745) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.42E+00 | Statistic Test | p-value: 4.34E-02; Z-score: 6.73E-01 | ||
|
Methylation in Case |
8.83E-01 (Median) | Methylation in Control | 6.22E-01 (Median) | ||
|
Studied Phenotype |
Panic disorder [ ICD-11: 6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC25A2 in panic disorder | [ 6 ] | |||
|
Location |
1stExon (cg03060468) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.70E+00 | Statistic Test | p-value: 1.19E-02; Z-score: 4.24E-01 | ||
|
Methylation in Case |
4.37E-01 (Median) | Methylation in Control | 2.57E-01 (Median) | ||
|
Studied Phenotype |
Panic disorder [ ICD-11: 6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Papillary thyroid cancer |
9 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC25A2 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
TSS1500 (cg26783353) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.03E+00 | Statistic Test | p-value: 1.92E-03; Z-score: 7.82E-01 | ||
|
Methylation in Case |
9.40E-01 (Median) | Methylation in Control | 9.17E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC25A2 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
TSS1500 (cg22748470) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.07E+00 | Statistic Test | p-value: 2.49E-03; Z-score: 8.65E-01 | ||
|
Methylation in Case |
7.46E-01 (Median) | Methylation in Control | 7.00E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC25A2 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
TSS200 (cg07496545) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.33E+00 | Statistic Test | p-value: 6.87E-06; Z-score: 1.22E+00 | ||
|
Methylation in Case |
4.38E-01 (Median) | Methylation in Control | 3.30E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC25A2 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
TSS200 (cg05845376) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.39E+00 | Statistic Test | p-value: 2.79E-05; Z-score: 9.96E-01 | ||
|
Methylation in Case |
4.13E-01 (Median) | Methylation in Control | 2.97E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC25A2 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
TSS200 (cg25212131) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.11E+00 | Statistic Test | p-value: 1.19E-04; Z-score: 9.33E-01 | ||
|
Methylation in Case |
5.28E-01 (Median) | Methylation in Control | 4.77E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC25A2 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
TSS200 (cg07039560) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.17E+00 | Statistic Test | p-value: 8.63E-04; Z-score: 1.06E+00 | ||
|
Methylation in Case |
3.85E-01 (Median) | Methylation in Control | 3.28E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC25A2 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
TSS200 (cg00030420) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.09E+00 | Statistic Test | p-value: 1.72E-03; Z-score: 9.32E-01 | ||
|
Methylation in Case |
7.26E-01 (Median) | Methylation in Control | 6.65E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC25A2 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
TSS200 (cg27320005) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.13E+00 | Statistic Test | p-value: 8.29E-03; Z-score: 6.91E-01 | ||
|
Methylation in Case |
5.08E-01 (Median) | Methylation in Control | 4.51E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLC25A2 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
1stExon (cg05638493) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.07E+00 | Statistic Test | p-value: 4.70E-03; Z-score: 2.66E-01 | ||
|
Methylation in Case |
2.32E-01 (Median) | Methylation in Control | 2.18E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Prostate cancer |
10 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC25A2 in prostate cancer | [ 8 ] | |||
|
Location |
TSS1500 (cg10726357) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.24E+00 | Statistic Test | p-value: 2.31E-03; Z-score: 4.57E+00 | ||
|
Methylation in Case |
8.69E-01 (Median) | Methylation in Control | 7.02E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC25A2 in prostate cancer | [ 8 ] | |||
|
Location |
TSS1500 (cg09110274) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.06E+00 | Statistic Test | p-value: 7.48E-03; Z-score: 3.11E+00 | ||
|
Methylation in Case |
9.08E-01 (Median) | Methylation in Control | 8.60E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC25A2 in prostate cancer | [ 8 ] | |||
|
Location |
TSS1500 (cg02505749) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 2.29E+00 | Statistic Test | p-value: 9.45E-03; Z-score: 1.78E+01 | ||
|
Methylation in Case |
1.99E-01 (Median) | Methylation in Control | 8.67E-02 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC25A2 in prostate cancer | [ 8 ] | |||
|
Location |
TSS200 (cg07578695) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 5.18E+00 | Statistic Test | p-value: 1.15E-02; Z-score: 1.98E+01 | ||
|
Methylation in Case |
3.63E-01 (Median) | Methylation in Control | 7.00E-02 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC25A2 in prostate cancer | [ 8 ] | |||
|
Location |
Body (cg14565651) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.39E+00 | Statistic Test | p-value: 2.05E-03; Z-score: 4.86E+00 | ||
|
Methylation in Case |
7.72E-01 (Median) | Methylation in Control | 5.57E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC25A2 in prostate cancer | [ 8 ] | |||
|
Location |
Body (cg18741958) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.38E+00 | Statistic Test | p-value: 2.25E-03; Z-score: 6.07E+00 | ||
|
Methylation in Case |
8.49E-01 (Median) | Methylation in Control | 6.14E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC25A2 in prostate cancer | [ 8 ] | |||
|
Location |
Body (cg05418487) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.45E+00 | Statistic Test | p-value: 3.01E-03; Z-score: 4.57E+00 | ||
|
Methylation in Case |
7.11E-01 (Median) | Methylation in Control | 4.90E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC25A2 in prostate cancer | [ 8 ] | |||
|
Location |
Body (cg08638180) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.84E+00 | Statistic Test | p-value: 5.49E-03; Z-score: 9.34E+00 | ||
|
Methylation in Case |
7.27E-01 (Median) | Methylation in Control | 3.96E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLC25A2 in prostate cancer | [ 8 ] | |||
|
Location |
Body (cg13058819) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.70E+00 | Statistic Test | p-value: 1.88E-02; Z-score: -2.32E+00 | ||
|
Methylation in Case |
2.41E-01 (Median) | Methylation in Control | 4.10E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 10 |
Moderate hypermethylation of SLC25A2 in prostate cancer than that in healthy individual | ||||
Studied Phenotype |
Prostate cancer [ICD-11:2C82] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 2.30E-05; Fold-change: 0.235872199; Z-score: 2.459161114 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
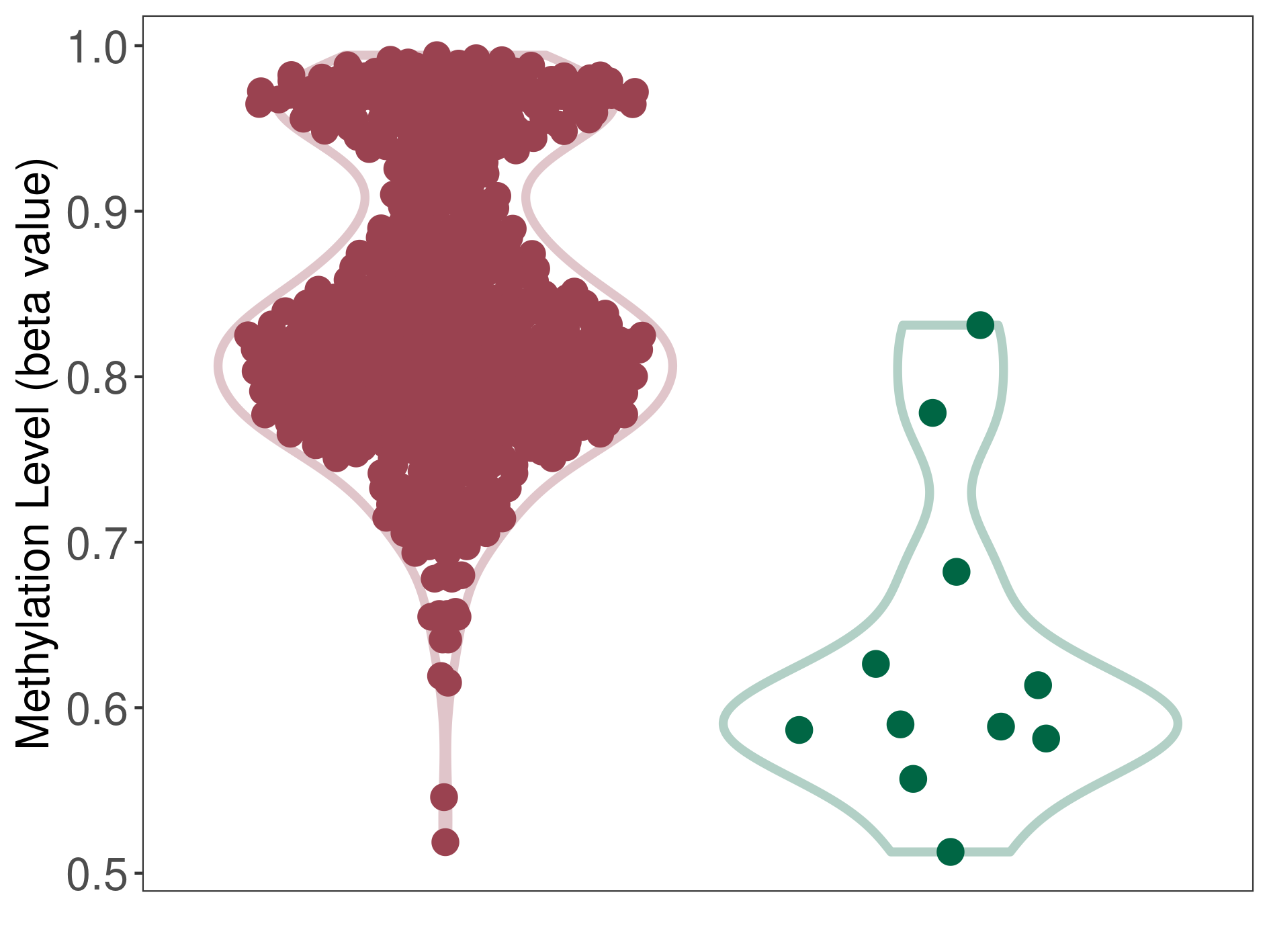
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Breast cancer |
9 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC25A2 in breast cancer | [ 9 ] | |||
|
Location |
TSS200 (cg21636683) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.33E+00 | Statistic Test | p-value: 1.45E-28; Z-score: 3.70E+00 | ||
|
Methylation in Case |
6.89E-01 (Median) | Methylation in Control | 5.18E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC25A2 in breast cancer | [ 9 ] | |||
|
Location |
TSS200 (cg05845376) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.68E+00 | Statistic Test | p-value: 1.11E-27; Z-score: 3.45E+00 | ||
|
Methylation in Case |
8.15E-01 (Median) | Methylation in Control | 4.85E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC25A2 in breast cancer | [ 9 ] | |||
|
Location |
TSS200 (cg07496545) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.60E+00 | Statistic Test | p-value: 1.29E-22; Z-score: 2.77E+00 | ||
|
Methylation in Case |
8.08E-01 (Median) | Methylation in Control | 5.04E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC25A2 in breast cancer | [ 9 ] | |||
|
Location |
TSS200 (cg00030420) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.13E+00 | Statistic Test | p-value: 5.93E-18; Z-score: 2.21E+00 | ||
|
Methylation in Case |
8.03E-01 (Median) | Methylation in Control | 7.10E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC25A2 in breast cancer | [ 9 ] | |||
|
Location |
TSS200 (cg27320005) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.38E+00 | Statistic Test | p-value: 5.02E-16; Z-score: 2.45E+00 | ||
|
Methylation in Case |
7.86E-01 (Median) | Methylation in Control | 5.70E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC25A2 in breast cancer | [ 9 ] | |||
|
Location |
TSS200 (cg25212131) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.25E+00 | Statistic Test | p-value: 2.15E-14; Z-score: 1.56E+00 | ||
|
Methylation in Case |
6.88E-01 (Median) | Methylation in Control | 5.49E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC25A2 in breast cancer | [ 9 ] | |||
|
Location |
TSS200 (cg07039560) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.44E+00 | Statistic Test | p-value: 1.71E-13; Z-score: 1.54E+00 | ||
|
Methylation in Case |
5.66E-01 (Median) | Methylation in Control | 3.92E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC25A2 in breast cancer | [ 9 ] | |||
|
Location |
1stExon (cg05638493) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.62E+00 | Statistic Test | p-value: 8.19E-27; Z-score: 3.86E+00 | ||
|
Methylation in Case |
5.46E-01 (Median) | Methylation in Control | 3.37E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLC25A2 in breast cancer | [ 9 ] | |||
|
Location |
1stExon (cg03060468) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.18E+00 | Statistic Test | p-value: 1.76E-11; Z-score: 1.66E+00 | ||
|
Methylation in Case |
6.02E-01 (Median) | Methylation in Control | 5.12E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
HIV infection |
10 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC25A2 in HIV infection | [ 10 ] | |||
|
Location |
TSS200 (cg00030420) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.07E+00 | Statistic Test | p-value: 4.61E-07; Z-score: 1.28E+00 | ||
|
Methylation in Case |
8.54E-01 (Median) | Methylation in Control | 7.97E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC25A2 in HIV infection | [ 10 ] | |||
|
Location |
TSS200 (cg07039560) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.15E+00 | Statistic Test | p-value: 8.47E-06; Z-score: 1.46E+00 | ||
|
Methylation in Case |
7.71E-01 (Median) | Methylation in Control | 6.68E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC25A2 in HIV infection | [ 10 ] | |||
|
Location |
TSS200 (cg27320005) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.10E+00 | Statistic Test | p-value: 9.15E-06; Z-score: 1.20E+00 | ||
|
Methylation in Case |
8.78E-01 (Median) | Methylation in Control | 8.02E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC25A2 in HIV infection | [ 10 ] | |||
|
Location |
TSS200 (cg25212131) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.08E+00 | Statistic Test | p-value: 1.89E-04; Z-score: 1.03E+00 | ||
|
Methylation in Case |
8.38E-01 (Median) | Methylation in Control | 7.77E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC25A2 in HIV infection | [ 10 ] | |||
|
Location |
TSS200 (cg07496545) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.05E+00 | Statistic Test | p-value: 2.95E-04; Z-score: 6.00E-01 | ||
|
Methylation in Case |
9.17E-01 (Median) | Methylation in Control | 8.74E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC25A2 in HIV infection | [ 10 ] | |||
|
Location |
TSS200 (cg21636683) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.05E+00 | Statistic Test | p-value: 4.22E-03; Z-score: 7.69E-01 | ||
|
Methylation in Case |
8.45E-01 (Median) | Methylation in Control | 8.04E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC25A2 in HIV infection | [ 10 ] | |||
|
Location |
TSS200 (cg05845376) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.07E+00 | Statistic Test | p-value: 5.29E-03; Z-score: 7.87E-01 | ||
|
Methylation in Case |
9.20E-01 (Median) | Methylation in Control | 8.56E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC25A2 in HIV infection | [ 10 ] | |||
|
Location |
1stExon (cg05638493) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.20E+00 | Statistic Test | p-value: 1.62E-05; Z-score: 1.55E+00 | ||
|
Methylation in Case |
5.24E-01 (Median) | Methylation in Control | 4.37E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLC25A2 in HIV infection | [ 10 ] | |||
|
Location |
1stExon (cg02553537) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.03E+00 | Statistic Test | p-value: 2.29E-03; Z-score: -7.32E-01 | ||
|
Methylation in Case |
8.73E-01 (Median) | Methylation in Control | 9.00E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 10 |
Methylation of SLC25A2 in HIV infection | [ 10 ] | |||
|
Location |
1stExon (cg03060468) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.10E+00 | Statistic Test | p-value: 9.45E-03; Z-score: 1.04E+00 | ||
|
Methylation in Case |
6.67E-01 (Median) | Methylation in Control | 6.04E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Lung adenocarcinoma |
9 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC25A2 in lung adenocarcinoma | [ 11 ] | |||
|
Location |
TSS200 (cg05845376) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.23E+00 | Statistic Test | p-value: 9.75E-05; Z-score: 2.23E+00 | ||
|
Methylation in Case |
8.50E-01 (Median) | Methylation in Control | 6.93E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC25A2 in lung adenocarcinoma | [ 11 ] | |||
|
Location |
TSS200 (cg07496545) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.19E+00 | Statistic Test | p-value: 1.01E-03; Z-score: 1.96E+00 | ||
|
Methylation in Case |
8.70E-01 (Median) | Methylation in Control | 7.34E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC25A2 in lung adenocarcinoma | [ 11 ] | |||
|
Location |
TSS200 (cg27320005) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.14E+00 | Statistic Test | p-value: 3.16E-03; Z-score: 1.58E+00 | ||
|
Methylation in Case |
8.44E-01 (Median) | Methylation in Control | 7.43E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC25A2 in lung adenocarcinoma | [ 11 ] | |||
|
Location |
TSS200 (cg00030420) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.09E+00 | Statistic Test | p-value: 3.45E-03; Z-score: 1.77E+00 | ||
|
Methylation in Case |
8.58E-01 (Median) | Methylation in Control | 7.84E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC25A2 in lung adenocarcinoma | [ 11 ] | |||
|
Location |
TSS200 (cg25212131) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.14E+00 | Statistic Test | p-value: 4.86E-03; Z-score: 1.76E+00 | ||
|
Methylation in Case |
7.71E-01 (Median) | Methylation in Control | 6.75E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC25A2 in lung adenocarcinoma | [ 11 ] | |||
|
Location |
TSS200 (cg07039560) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.20E+00 | Statistic Test | p-value: 8.86E-03; Z-score: 1.69E+00 | ||
|
Methylation in Case |
7.05E-01 (Median) | Methylation in Control | 5.87E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC25A2 in lung adenocarcinoma | [ 11 ] | |||
|
Location |
TSS200 (cg21636683) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.12E+00 | Statistic Test | p-value: 1.44E-02; Z-score: 1.53E+00 | ||
|
Methylation in Case |
7.77E-01 (Median) | Methylation in Control | 6.92E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC25A2 in lung adenocarcinoma | [ 11 ] | |||
|
Location |
1stExon (cg05638493) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.38E+00 | Statistic Test | p-value: 3.30E-04; Z-score: 2.55E+00 | ||
|
Methylation in Case |
5.96E-01 (Median) | Methylation in Control | 4.30E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLC25A2 in lung adenocarcinoma | [ 11 ] | |||
|
Location |
1stExon (cg03060468) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.16E+00 | Statistic Test | p-value: 1.00E-02; Z-score: 1.70E+00 | ||
|
Methylation in Case |
6.71E-01 (Median) | Methylation in Control | 5.80E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Pancretic ductal adenocarcinoma |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC25A2 in pancretic ductal adenocarcinoma | [ 12 ] | |||
|
Location |
TSS200 (cg06956052) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.02E+00 | Statistic Test | p-value: 2.97E-02; Z-score: -3.88E-01 | ||
|
Methylation in Case |
5.16E-01 (Median) | Methylation in Control | 5.29E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC25A2 in pancretic ductal adenocarcinoma | [ 12 ] | |||
|
Location |
Body (cg19639560) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.05E+00 | Statistic Test | p-value: 3.50E-02; Z-score: 3.15E-01 | ||
|
Methylation in Case |
6.94E-01 (Median) | Methylation in Control | 6.64E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC25A2 in pancretic ductal adenocarcinoma | [ 12 ] | |||
|
Location |
Body (cg10752421) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.01E+00 | Statistic Test | p-value: 4.83E-02; Z-score: 3.96E-01 | ||
|
Methylation in Case |
9.31E-01 (Median) | Methylation in Control | 9.20E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Systemic lupus erythematosus |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC25A2 in systemic lupus erythematosus | [ 13 ] | |||
|
Location |
TSS200 (cg07496545) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.03E+00 | Statistic Test | p-value: 1.48E-02; Z-score: 2.47E-01 | ||
|
Methylation in Case |
8.81E-01 (Median) | Methylation in Control | 8.58E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus [ ICD-11: 4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC25A2 in systemic lupus erythematosus | [ 13 ] | |||
|
Location |
TSS200 (cg00030420) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.01E+00 | Statistic Test | p-value: 3.05E-02; Z-score: 8.83E-02 | ||
|
Methylation in Case |
8.65E-01 (Median) | Methylation in Control | 8.58E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus [ ICD-11: 4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Atypical teratoid rhabdoid tumor |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC25A2 in atypical teratoid rhabdoid tumor | [ 14 ] | |||
|
Location |
1stExon (cg02553537) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.39E+00 | Statistic Test | p-value: 4.27E-27; Z-score: -4.87E+00 | ||
|
Methylation in Case |
5.76E-01 (Median) | Methylation in Control | 8.03E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC25A2 in atypical teratoid rhabdoid tumor | [ 14 ] | |||
|
Location |
1stExon (cg03060468) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 2.62E+00 | Statistic Test | p-value: 6.72E-26; Z-score: 5.33E+00 | ||
|
Methylation in Case |
3.72E-01 (Median) | Methylation in Control | 1.42E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC25A2 in atypical teratoid rhabdoid tumor | [ 14 ] | |||
|
Location |
1stExon (cg05638493) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.41E+00 | Statistic Test | p-value: 1.18E-23; Z-score: -5.03E+00 | ||
|
Methylation in Case |
5.98E-01 (Median) | Methylation in Control | 8.45E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Anaplastic pleomorphic xanthoastrocytoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Moderate hypermethylation of SLC25A2 in anaplastic pleomorphic xanthoastrocytoma than that in healthy individual | ||||
Studied Phenotype |
Anaplastic pleomorphic xanthoastrocytoma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 2.99E-05; Fold-change: 0.260927344; Z-score: 1.165572732 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
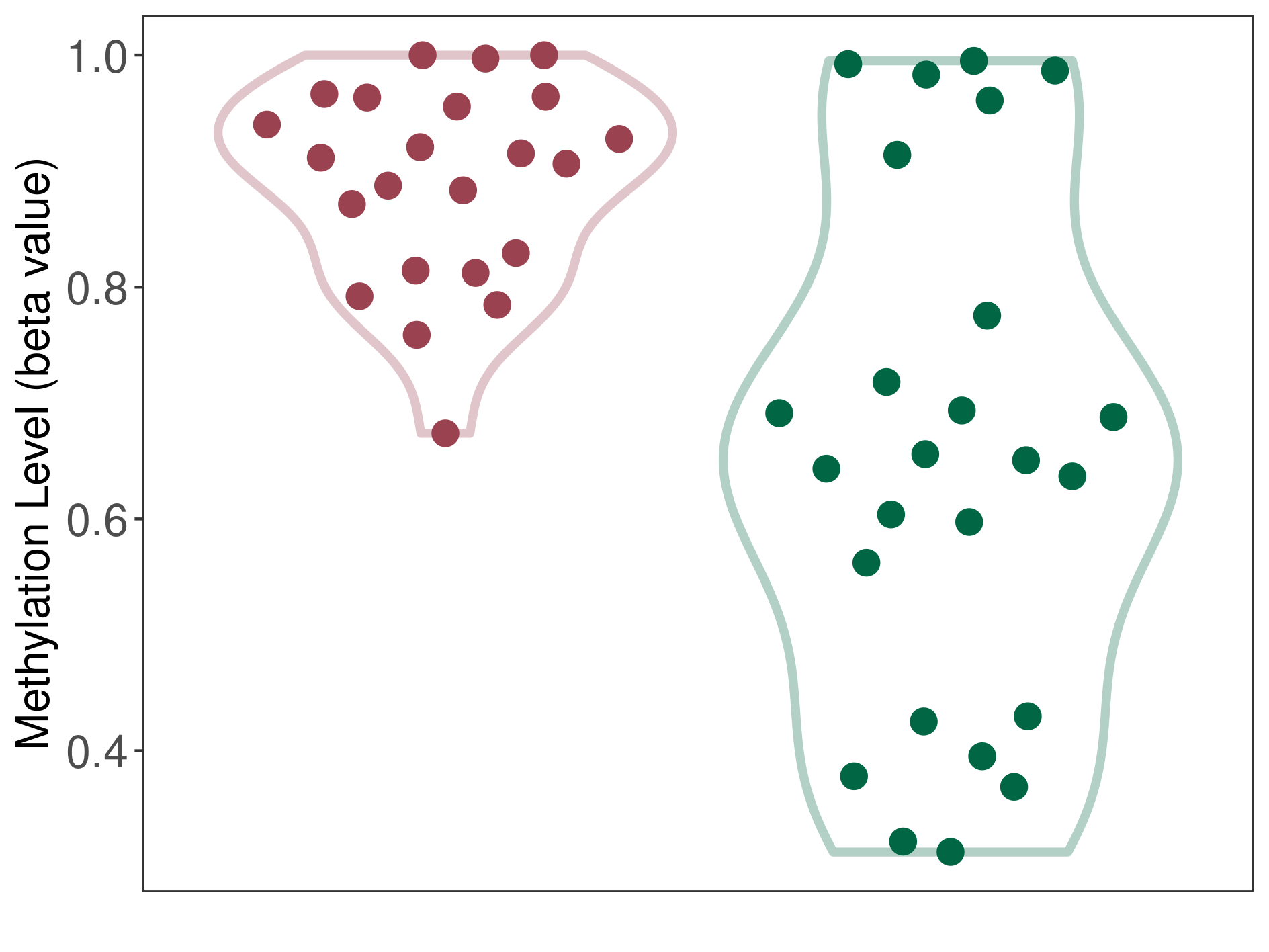
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Atypical teratoid rhabdoid tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Moderate hypermethylation of SLC25A2 in atypical teratoid rhabdoid tumour than that in healthy individual | ||||
Studied Phenotype |
Atypical teratoid rhabdoid tumour [ICD-11:2A00.1Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 2.74E-09; Fold-change: 0.254327024; Z-score: 1.279206047 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||

|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Brain neuroblastoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Moderate hypermethylation of SLC25A2 in brain neuroblastoma than that in healthy individual | ||||
Studied Phenotype |
Brain neuroblastoma [ICD-11:2A00.11] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 5.84E-09; Fold-change: 0.28534596; Z-score: 1.497684076 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||

|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Brain neuroepithelial tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Moderate hypermethylation of SLC25A2 in brain neuroepithelial tumour than that in healthy individual | ||||
Studied Phenotype |
Brain neuroepithelial tumour [ICD-11:2A00.2Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 3.20E-12; Fold-change: 0.222589169; Z-score: 1.32162559 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||

|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Medulloblastoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Moderate hypermethylation of SLC25A2 in medulloblastoma than that in healthy individual | ||||
Studied Phenotype |
Medulloblastoma [ICD-11:2A00.10] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 9.56E-25; Fold-change: 0.236984055; Z-score: 1.786196764 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
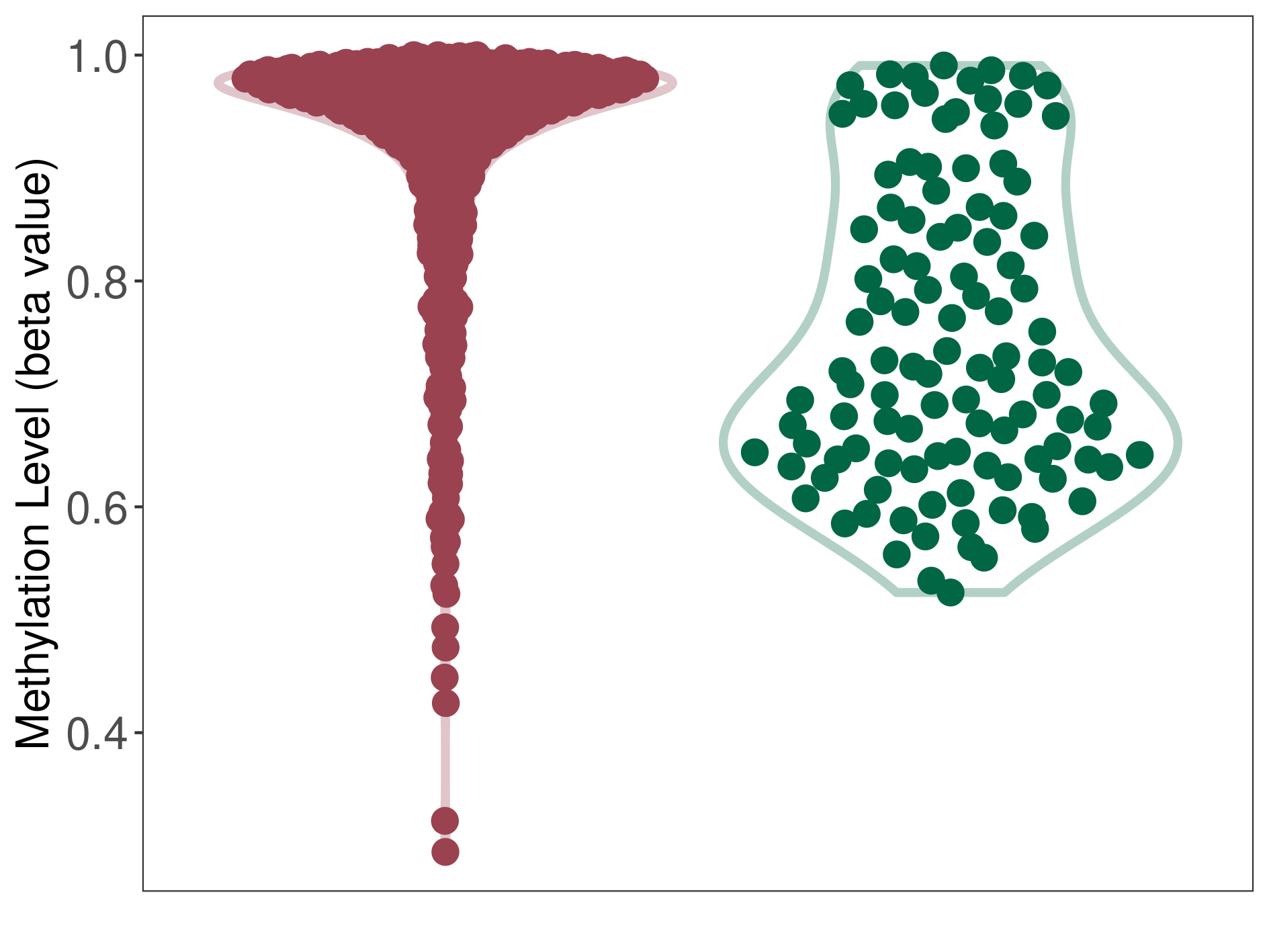
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Melanoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Moderate hypermethylation of SLC25A2 in melanoma than that in healthy individual | ||||
Studied Phenotype |
Melanoma [ICD-11:2C30] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 8.86E-13; Fold-change: 0.274282057; Z-score: 1.56910286 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
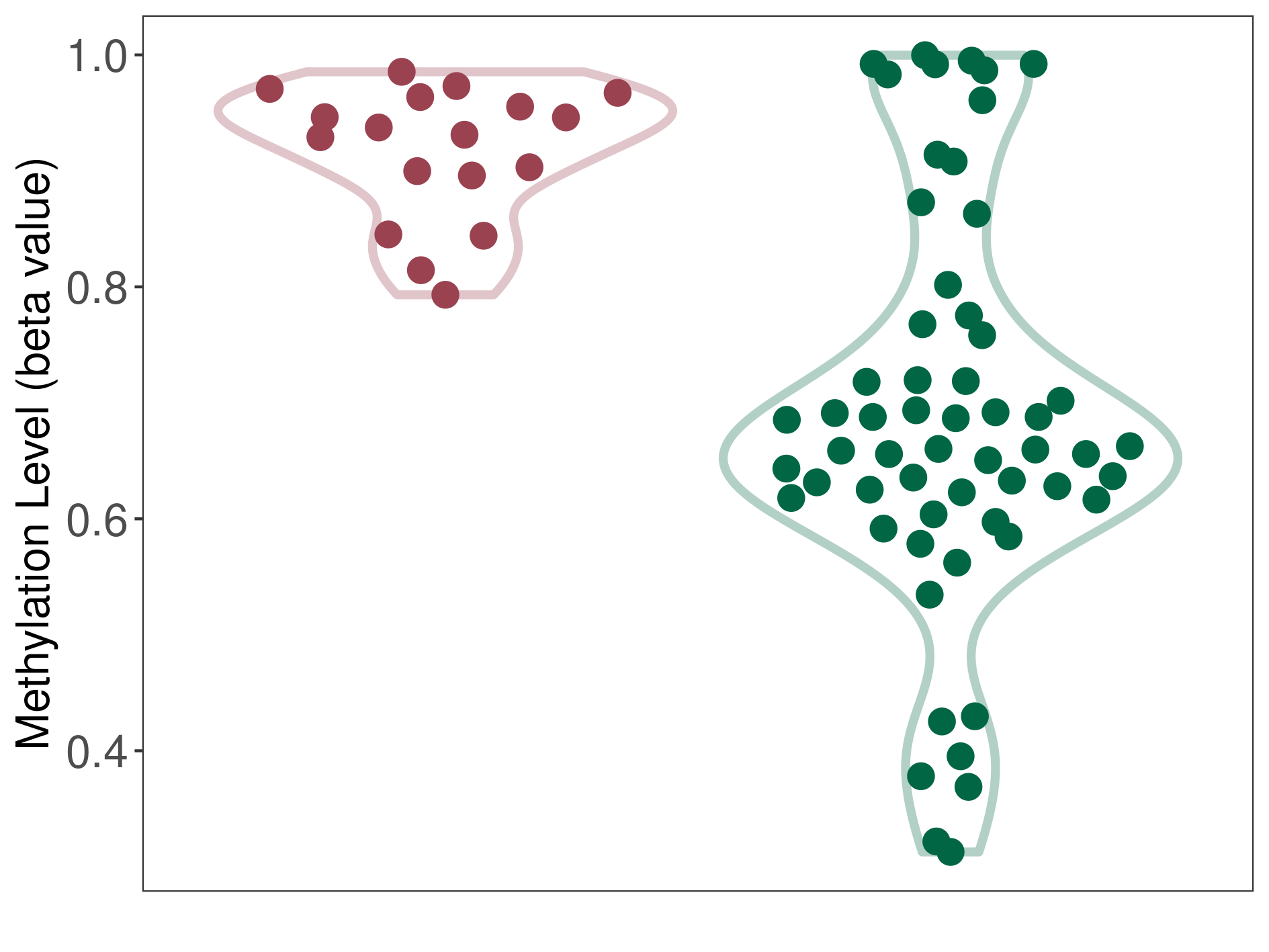
|
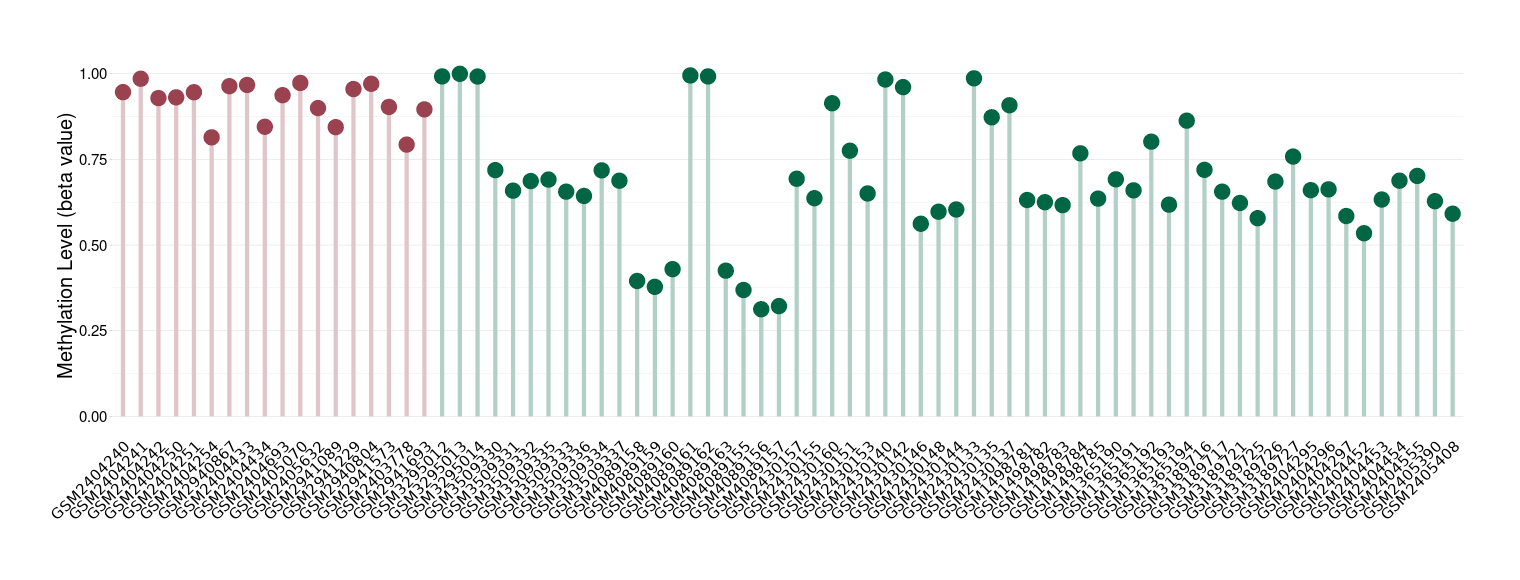 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Peripheral neuroectodermal tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Moderate hypermethylation of SLC25A2 in peripheral neuroectodermal tumour than that in healthy individual | ||||
Studied Phenotype |
Peripheral neuroectodermal tumour [ICD-11:2B52] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 1.84E-05; Fold-change: 0.230117501; Z-score: 1.108801957 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||

|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Prostate cancer metastasis |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Moderate hypermethylation of SLC25A2 in prostate cancer metastasis than that in healthy individual | ||||
Studied Phenotype |
Prostate cancer metastasis [ICD-11:2.00E+06] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 1.83E-05; Fold-change: 0.260209701; Z-score: 6.195582292 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
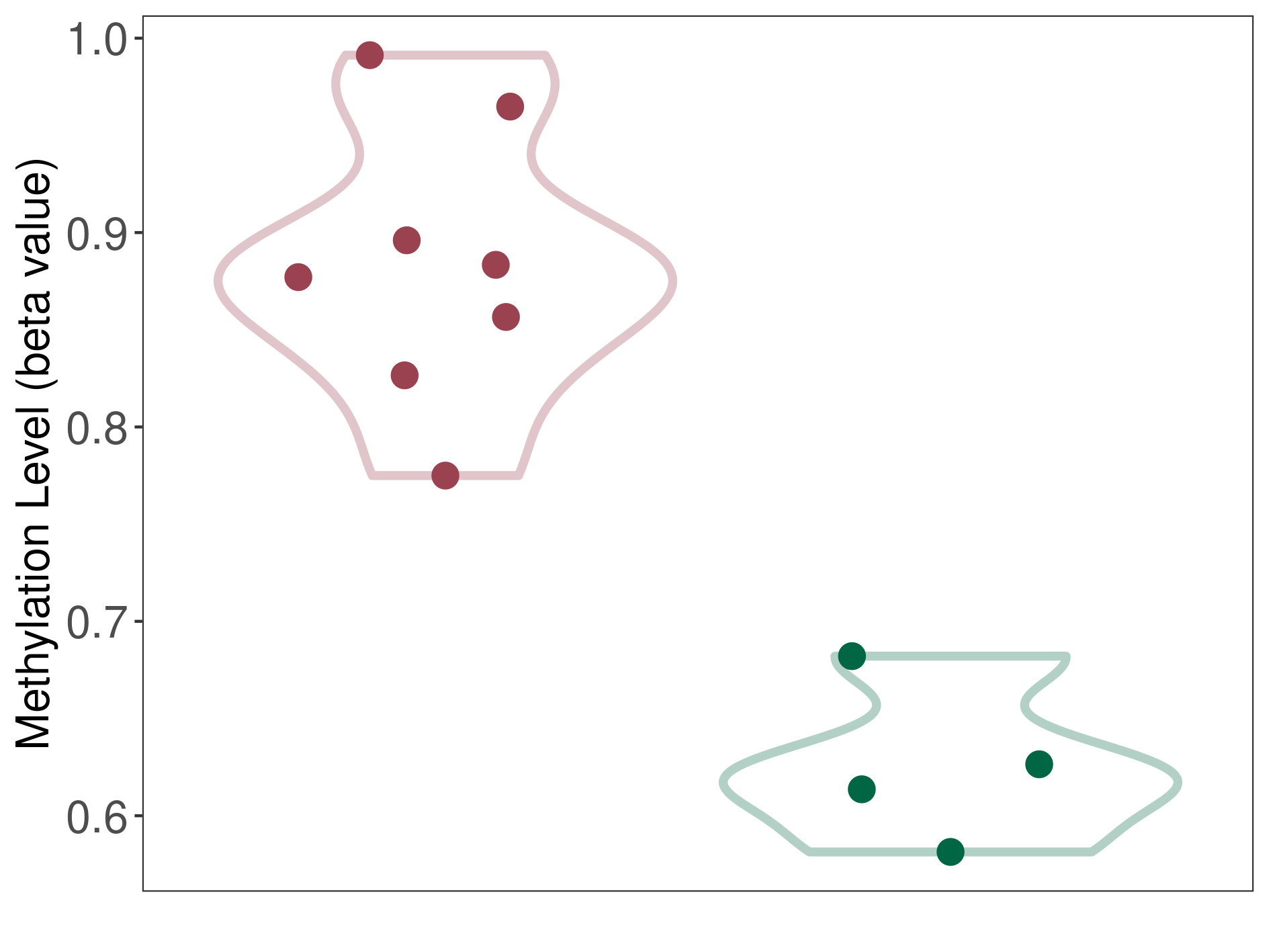
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
RELA YAP fusion ependymoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Moderate hypermethylation of SLC25A2 in rela yap fusion ependymoma than that in healthy individual | ||||
Studied Phenotype |
RELA YAP fusion ependymoma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 2.05E-18; Fold-change: 0.235358029; Z-score: 1.599491962 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||

|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Third ventricle chordoid glioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Moderate hypermethylation of SLC25A2 in third ventricle chordoid glioma than that in healthy individual | ||||
Studied Phenotype |
Third ventricle chordoid glioma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 0.005457792; Fold-change: 0.240104097; Z-score: 1.065850813 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||

|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Pancreatic cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Moderate hypomethylation of SLC25A2 in pancreatic adenocarcinoma than that in healthy individual | ||||
Studied Phenotype |
Pancreatic cancer [ICD-11:2C10] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 0.034901044; Fold-change: -0.224297589; Z-score: -3.544077934 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue adjacent to the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
Multilayered rosettes embryonal tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypermethylation of SLC25A2 in multilayered rosettes embryonal tumour than that in healthy individual | ||||
Studied Phenotype |
Multilayered rosettes embryonal tumour [ICD-11:2A00.1] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 1.34E-11; Fold-change: 0.313091048; Z-score: 1.663505199 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||

|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Uterine carcinosarcoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypermethylation of SLC25A2 in uterine carcinosarcoma than that in healthy individual | ||||
Studied Phenotype |
Uterine carcinosarcoma [ICD-11:2B5F] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 3.37E-06; Fold-change: 0.304275576; Z-score: 2.815067823 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
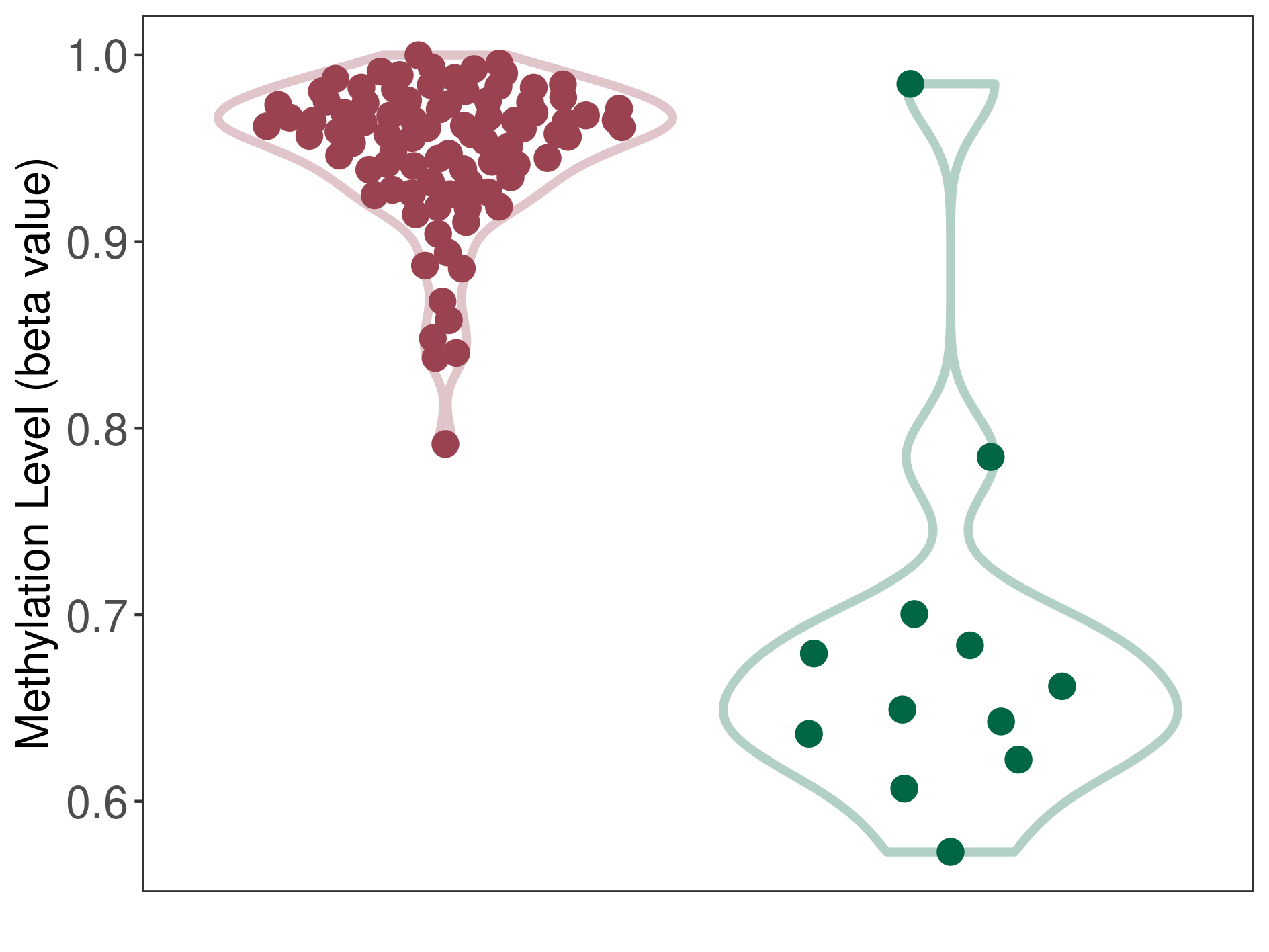
|
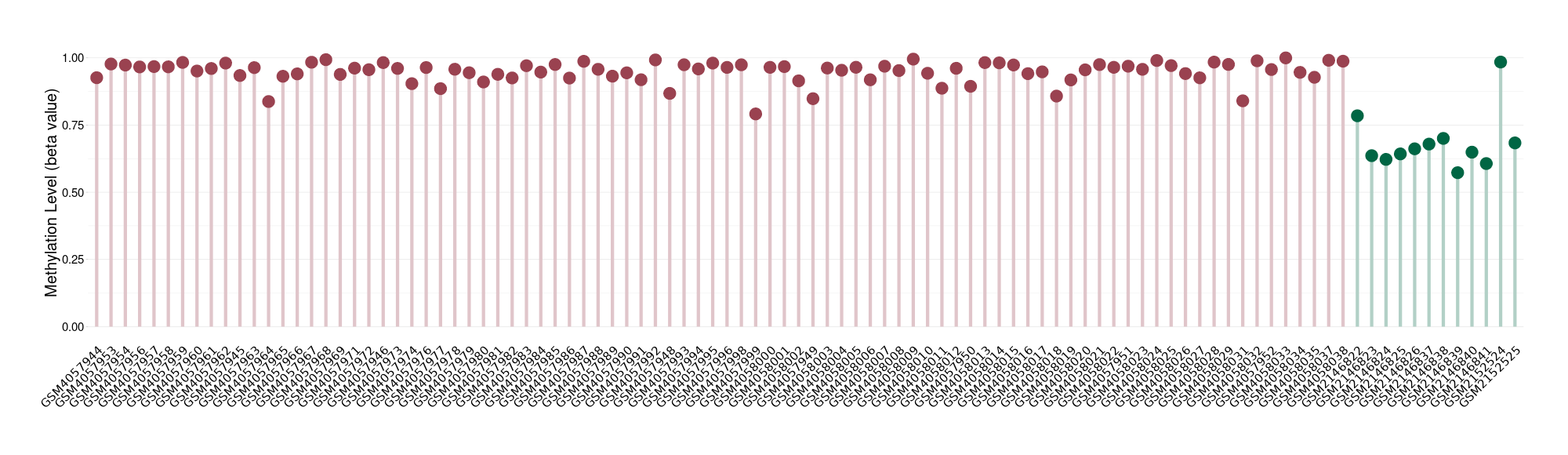 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Obesity |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypomethylation of SLC25A2 in obesity than that in healthy individual | ||||
Studied Phenotype |
Obesity [ICD-11:5B81] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 1.02E-32; Fold-change: -0.387124081; Z-score: -3.248816818 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
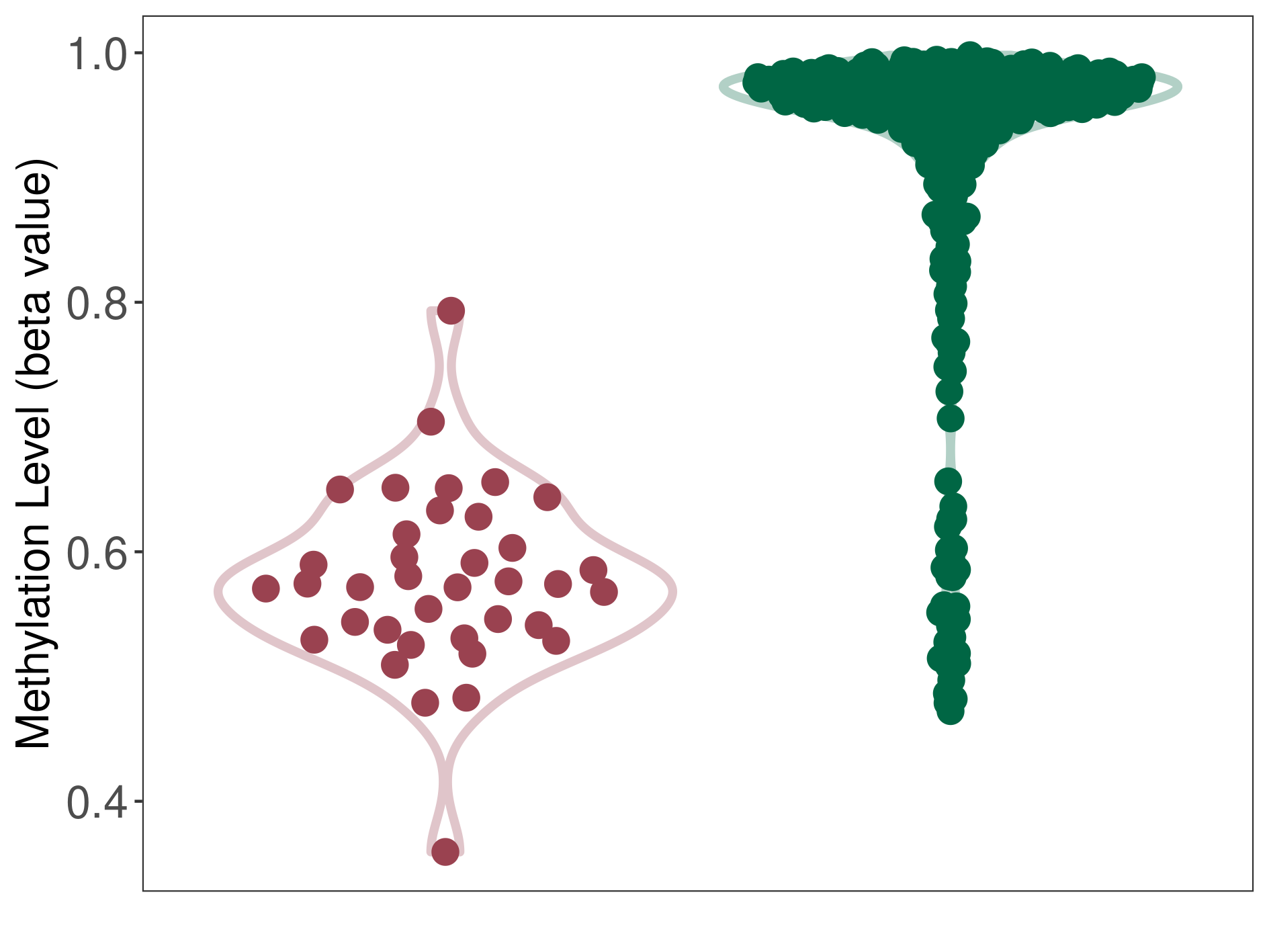
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
microRNA |
|||||
|
Unclear Phenotype |
9 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
miR-3190 directly targets SLC25A2 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-3190 | miRNA Mature ID | miR-3190-5p | ||
|
miRNA Sequence |
UCUGGCCAGCUACGUCCCCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 2 |
miR-378a directly targets SLC25A2 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | CLASH | ||
|
miRNA Stemloop ID |
miR-378a | miRNA Mature ID | miR-378a-3p | ||
|
miRNA Sequence |
ACUGGACUUGGAGUCAGAAGGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon 3 |
miR-3926 directly targets SLC25A2 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-3926 | miRNA Mature ID | miR-3926 | ||
|
miRNA Sequence |
UGGCCAAAAAGCAGGCAGAGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 4 |
miR-4786 directly targets SLC25A2 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4786 | miRNA Mature ID | miR-4786-5p | ||
|
miRNA Sequence |
UGAGACCAGGACUGGAUGCACC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 5 |
miR-548c directly targets SLC25A2 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-548c | miRNA Mature ID | miR-548c-3p | ||
|
miRNA Sequence |
CAAAAAUCUCAAUUACUUUUGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 6 |
miR-548s directly targets SLC25A2 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-548s | miRNA Mature ID | miR-548s | ||
|
miRNA Sequence |
AUGGCCAAAACUGCAGUUAUUUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 7 |
miR-6728 directly targets SLC25A2 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6728 | miRNA Mature ID | miR-6728-3p | ||
|
miRNA Sequence |
UCUCUGCUCUGCUCUCCCCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 8 |
miR-6771 directly targets SLC25A2 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6771 | miRNA Mature ID | miR-6771-3p | ||
|
miRNA Sequence |
CAAACCCCUGUCUACCCGCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 9 |
miR-6849 directly targets SLC25A2 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6849 | miRNA Mature ID | miR-6849-3p | ||
|
miRNA Sequence |
ACCAGCCUGUGUCCACCUCCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.
