Detail Information of Epigenetic Regulations
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0425 Transporter Info | ||||
| Gene Name | SLC5A5 | ||||
| Transporter Name | Sodium/iodide cotransporter | ||||
| Gene ID | |||||
| UniProt ID | |||||
| Epigenetic Regulations of This DT (EGR) | |||||
|---|---|---|---|---|---|
|
Methylation |
|||||
|
Pancretic ductal adenocarcinoma |
9 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC5A5 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
5'UTR (cg08140412) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.36E+00 | Statistic Test | p-value: 1.10E-25; Z-score: -3.98E+00 | ||
|
Methylation in Case |
3.11E-01 (Median) | Methylation in Control | 4.24E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC5A5 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg09028651) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.10E+00 | Statistic Test | p-value: 2.05E-03; Z-score: 7.31E-01 | ||
|
Methylation in Case |
7.49E-01 (Median) | Methylation in Control | 6.83E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC5A5 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg07496545) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.49E+00 | Statistic Test | p-value: 1.14E-06; Z-score: 1.58E+00 | ||
|
Methylation in Case |
7.41E-01 (Median) | Methylation in Control | 4.97E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC5A5 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
1stExon (cg09597465) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.10E+00 | Statistic Test | p-value: 1.69E-04; Z-score: -6.19E-01 | ||
|
Methylation in Case |
9.68E-02 (Median) | Methylation in Control | 1.06E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC5A5 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg12178904) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.29E+00 | Statistic Test | p-value: 2.84E-26; Z-score: -5.31E+00 | ||
|
Methylation in Case |
6.08E-01 (Median) | Methylation in Control | 7.83E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC5A5 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg02685680) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.34E+00 | Statistic Test | p-value: 6.96E-08; Z-score: -1.73E+00 | ||
|
Methylation in Case |
6.12E-01 (Median) | Methylation in Control | 8.20E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC5A5 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg14014879) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.17E+00 | Statistic Test | p-value: 1.10E-07; Z-score: -1.67E+00 | ||
|
Methylation in Case |
6.20E-01 (Median) | Methylation in Control | 7.26E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC5A5 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg09725874) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.12E+00 | Statistic Test | p-value: 5.66E-04; Z-score: -1.12E+00 | ||
|
Methylation in Case |
3.15E-01 (Median) | Methylation in Control | 3.54E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLC5A5 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg21271026) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.05E+00 | Statistic Test | p-value: 1.41E-03; Z-score: 9.13E-01 | ||
|
Methylation in Case |
8.30E-01 (Median) | Methylation in Control | 7.93E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Bladder cancer |
9 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC5A5 in bladder cancer | [ 2 ] | |||
|
Location |
TSS1500 (cg10017118) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.34E+00 | Statistic Test | p-value: 1.08E-06; Z-score: -1.04E+01 | ||
|
Methylation in Case |
3.94E-01 (Median) | Methylation in Control | 5.26E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC5A5 in bladder cancer | [ 2 ] | |||
|
Location |
TSS1500 (cg17933226) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.29E+00 | Statistic Test | p-value: 3.70E-03; Z-score: -6.05E+00 | ||
|
Methylation in Case |
4.21E-01 (Median) | Methylation in Control | 5.43E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC5A5 in bladder cancer | [ 2 ] | |||
|
Location |
TSS200 (cg16701007) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.55E+00 | Statistic Test | p-value: 5.23E-06; Z-score: -4.90E+00 | ||
|
Methylation in Case |
2.09E-01 (Median) | Methylation in Control | 3.22E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC5A5 in bladder cancer | [ 2 ] | |||
|
Location |
TSS200 (cg21477717) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.35E+00 | Statistic Test | p-value: 4.99E-03; Z-score: -3.04E+00 | ||
|
Methylation in Case |
1.13E-01 (Median) | Methylation in Control | 1.53E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC5A5 in bladder cancer | [ 2 ] | |||
|
Location |
Body (cg22179256) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.29E+00 | Statistic Test | p-value: 1.84E-06; Z-score: -1.39E+01 | ||
|
Methylation in Case |
6.26E-01 (Median) | Methylation in Control | 8.09E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC5A5 in bladder cancer | [ 2 ] | |||
|
Location |
Body (cg21522295) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.33E+00 | Statistic Test | p-value: 1.78E-04; Z-score: -5.03E+00 | ||
|
Methylation in Case |
1.19E-01 (Median) | Methylation in Control | 1.59E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC5A5 in bladder cancer | [ 2 ] | |||
|
Location |
Body (cg15427234) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 2.28E+00 | Statistic Test | p-value: 3.64E-04; Z-score: 6.13E+01 | ||
|
Methylation in Case |
2.96E-01 (Median) | Methylation in Control | 1.29E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC5A5 in bladder cancer | [ 2 ] | |||
|
Location |
Body (cg10359333) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.03E+00 | Statistic Test | p-value: 5.37E-04; Z-score: -2.07E+01 | ||
|
Methylation in Case |
8.90E-01 (Median) | Methylation in Control | 9.14E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLC5A5 in bladder cancer | [ 2 ] | |||
|
Location |
3'UTR (cg12562245) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.17E+00 | Statistic Test | p-value: 5.61E-04; Z-score: -5.89E+00 | ||
|
Methylation in Case |
6.33E-01 (Median) | Methylation in Control | 7.42E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Breast cancer |
15 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC5A5 in breast cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg17933226) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.21E+00 | Statistic Test | p-value: 4.05E-05; Z-score: 1.08E+00 | ||
|
Methylation in Case |
5.31E-01 (Median) | Methylation in Control | 4.40E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC5A5 in breast cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg10017118) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.07E+00 | Statistic Test | p-value: 1.87E-03; Z-score: 9.21E-01 | ||
|
Methylation in Case |
5.22E-01 (Median) | Methylation in Control | 4.87E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC5A5 in breast cancer | [ 3 ] | |||
|
Location |
TSS200 (cg23222039) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.73E+00 | Statistic Test | p-value: 6.40E-13; Z-score: 3.52E+00 | ||
|
Methylation in Case |
1.64E-01 (Median) | Methylation in Control | 9.47E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC5A5 in breast cancer | [ 3 ] | |||
|
Location |
TSS200 (cg21477717) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.52E+00 | Statistic Test | p-value: 1.31E-10; Z-score: 2.82E+00 | ||
|
Methylation in Case |
1.96E-01 (Median) | Methylation in Control | 1.29E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC5A5 in breast cancer | [ 3 ] | |||
|
Location |
TSS200 (cg24065770) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.94E+00 | Statistic Test | p-value: 2.49E-08; Z-score: 1.66E+00 | ||
|
Methylation in Case |
3.48E-02 (Median) | Methylation in Control | 1.80E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC5A5 in breast cancer | [ 3 ] | |||
|
Location |
TSS200 (cg16701007) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.30E+00 | Statistic Test | p-value: 3.18E-08; Z-score: 2.74E+00 | ||
|
Methylation in Case |
4.05E-01 (Median) | Methylation in Control | 3.13E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC5A5 in breast cancer | [ 3 ] | |||
|
Location |
TSS200 (cg06697339) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 2.51E+00 | Statistic Test | p-value: 1.20E-07; Z-score: 3.12E+00 | ||
|
Methylation in Case |
7.10E-02 (Median) | Methylation in Control | 2.83E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC5A5 in breast cancer | [ 3 ] | |||
|
Location |
TSS200 (cg01655355) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.21E+00 | Statistic Test | p-value: 1.37E-04; Z-score: 6.09E-01 | ||
|
Methylation in Case |
5.45E-02 (Median) | Methylation in Control | 4.49E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLC5A5 in breast cancer | [ 3 ] | |||
|
Location |
TSS200 (cg02674588) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.12E+00 | Statistic Test | p-value: 7.48E-03; Z-score: 4.29E-01 | ||
|
Methylation in Case |
8.08E-02 (Median) | Methylation in Control | 7.21E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 10 |
Methylation of SLC5A5 in breast cancer | [ 3 ] | |||
|
Location |
1stExon (cg24881657) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.99E+00 | Statistic Test | p-value: 8.43E-15; Z-score: 4.54E+00 | ||
|
Methylation in Case |
2.58E-01 (Median) | Methylation in Control | 1.30E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 11 |
Methylation of SLC5A5 in breast cancer | [ 3 ] | |||
|
Location |
Body (cg21522295) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.43E+00 | Statistic Test | p-value: 3.55E-08; Z-score: 2.36E+00 | ||
|
Methylation in Case |
2.07E-01 (Median) | Methylation in Control | 1.45E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 12 |
Methylation of SLC5A5 in breast cancer | [ 3 ] | |||
|
Location |
Body (cg15427234) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.30E+00 | Statistic Test | p-value: 1.11E-07; Z-score: 1.41E+00 | ||
|
Methylation in Case |
1.88E-01 (Median) | Methylation in Control | 1.44E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 13 |
Methylation of SLC5A5 in breast cancer | [ 3 ] | |||
|
Location |
Body (cg10660916) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.19E+00 | Statistic Test | p-value: 3.47E-05; Z-score: 6.25E-01 | ||
|
Methylation in Case |
2.81E-01 (Median) | Methylation in Control | 2.37E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 14 |
Methylation of SLC5A5 in breast cancer | [ 3 ] | |||
|
Location |
Body (cg11340363) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.40E+00 | Statistic Test | p-value: 3.61E-03; Z-score: 7.66E-01 | ||
|
Methylation in Case |
2.35E-02 (Median) | Methylation in Control | 1.67E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 15 |
Methylation of SLC5A5 in breast cancer | [ 3 ] | |||
|
Location |
3'UTR (cg12562245) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.09E+00 | Statistic Test | p-value: 1.00E-02; Z-score: -8.22E-01 | ||
|
Methylation in Case |
6.76E-01 (Median) | Methylation in Control | 7.36E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Renal cell carcinoma |
12 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC5A5 in clear cell renal cell carcinoma | [ 4 ] | |||
|
Location |
TSS1500 (cg17933226) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.25E+00 | Statistic Test | p-value: 2.90E-05; Z-score: 1.70E+00 | ||
|
Methylation in Case |
6.65E-01 (Median) | Methylation in Control | 5.34E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC5A5 in clear cell renal cell carcinoma | [ 4 ] | |||
|
Location |
TSS200 (cg06697339) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.35E+00 | Statistic Test | p-value: 1.97E-05; Z-score: 2.13E+00 | ||
|
Methylation in Case |
3.06E-02 (Median) | Methylation in Control | 2.27E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC5A5 in clear cell renal cell carcinoma | [ 4 ] | |||
|
Location |
TSS200 (cg01655355) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.45E+00 | Statistic Test | p-value: 3.70E-05; Z-score: 1.71E+00 | ||
|
Methylation in Case |
1.90E-02 (Median) | Methylation in Control | 1.31E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC5A5 in clear cell renal cell carcinoma | [ 4 ] | |||
|
Location |
TSS200 (cg23222039) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.46E+00 | Statistic Test | p-value: 9.94E-05; Z-score: 1.65E+00 | ||
|
Methylation in Case |
7.97E-02 (Median) | Methylation in Control | 5.46E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC5A5 in clear cell renal cell carcinoma | [ 4 ] | |||
|
Location |
TSS200 (cg02674588) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.22E+00 | Statistic Test | p-value: 1.68E-04; Z-score: 1.21E+00 | ||
|
Methylation in Case |
4.20E-02 (Median) | Methylation in Control | 3.45E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC5A5 in clear cell renal cell carcinoma | [ 4 ] | |||
|
Location |
TSS200 (cg24065770) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.17E+00 | Statistic Test | p-value: 2.72E-03; Z-score: 9.75E-01 | ||
|
Methylation in Case |
2.15E-02 (Median) | Methylation in Control | 1.83E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC5A5 in clear cell renal cell carcinoma | [ 4 ] | |||
|
Location |
TSS200 (cg21477717) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.14E+00 | Statistic Test | p-value: 6.24E-03; Z-score: 5.70E-01 | ||
|
Methylation in Case |
1.23E-01 (Median) | Methylation in Control | 1.08E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC5A5 in clear cell renal cell carcinoma | [ 4 ] | |||
|
Location |
1stExon (cg24881657) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.21E+00 | Statistic Test | p-value: 2.79E-04; Z-score: 7.85E-01 | ||
|
Methylation in Case |
1.19E-01 (Median) | Methylation in Control | 9.81E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLC5A5 in clear cell renal cell carcinoma | [ 4 ] | |||
|
Location |
Body (cg10660916) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.25E+00 | Statistic Test | p-value: 2.24E-05; Z-score: 2.20E+00 | ||
|
Methylation in Case |
3.77E-01 (Median) | Methylation in Control | 3.02E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 10 |
Methylation of SLC5A5 in clear cell renal cell carcinoma | [ 4 ] | |||
|
Location |
Body (cg15427234) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.43E+00 | Statistic Test | p-value: 3.80E-05; Z-score: 2.37E+00 | ||
|
Methylation in Case |
1.80E-01 (Median) | Methylation in Control | 1.26E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 11 |
Methylation of SLC5A5 in clear cell renal cell carcinoma | [ 4 ] | |||
|
Location |
Body (cg11340363) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.03E+00 | Statistic Test | p-value: 1.29E-02; Z-score: 2.28E-01 | ||
|
Methylation in Case |
3.72E-02 (Median) | Methylation in Control | 3.61E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 12 |
Methylation of SLC5A5 in clear cell renal cell carcinoma | [ 4 ] | |||
|
Location |
Body (cg12122403) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.14E+00 | Statistic Test | p-value: 1.38E-02; Z-score: 7.32E-01 | ||
|
Methylation in Case |
2.91E-02 (Median) | Methylation in Control | 2.54E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colon cancer |
12 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC5A5 in colon adenocarcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg16678467) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.19E+00 | Statistic Test | p-value: 1.20E-05; Z-score: -3.53E+00 | ||
|
Methylation in Case |
6.81E-01 (Median) | Methylation in Control | 8.07E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC5A5 in colon adenocarcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg21861233) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.15E+00 | Statistic Test | p-value: 1.26E-05; Z-score: -2.72E+00 | ||
|
Methylation in Case |
6.43E-01 (Median) | Methylation in Control | 7.39E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC5A5 in colon adenocarcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg03273715) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.20E+00 | Statistic Test | p-value: 1.74E-05; Z-score: -3.52E+00 | ||
|
Methylation in Case |
5.02E-01 (Median) | Methylation in Control | 6.03E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC5A5 in colon adenocarcinoma | [ 5 ] | |||
|
Location |
TSS200 (cg07769015) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.32E+00 | Statistic Test | p-value: 4.77E-05; Z-score: -1.98E+00 | ||
|
Methylation in Case |
3.69E-01 (Median) | Methylation in Control | 4.88E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC5A5 in colon adenocarcinoma | [ 5 ] | |||
|
Location |
TSS200 (cg07191594) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.22E+00 | Statistic Test | p-value: 7.19E-05; Z-score: 1.70E+00 | ||
|
Methylation in Case |
4.95E-01 (Median) | Methylation in Control | 4.05E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC5A5 in colon adenocarcinoma | [ 5 ] | |||
|
Location |
Body (cg27470827) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.27E+00 | Statistic Test | p-value: 1.45E-05; Z-score: -1.91E+00 | ||
|
Methylation in Case |
4.58E-01 (Median) | Methylation in Control | 5.83E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC5A5 in colon adenocarcinoma | [ 5 ] | |||
|
Location |
Body (cg01420001) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.20E+00 | Statistic Test | p-value: 1.80E-05; Z-score: -3.22E+00 | ||
|
Methylation in Case |
6.02E-01 (Median) | Methylation in Control | 7.20E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC5A5 in colon adenocarcinoma | [ 5 ] | |||
|
Location |
Body (cg15867963) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.40E+00 | Statistic Test | p-value: 3.09E-05; Z-score: 2.30E+00 | ||
|
Methylation in Case |
5.89E-01 (Median) | Methylation in Control | 4.21E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLC5A5 in colon adenocarcinoma | [ 5 ] | |||
|
Location |
Body (cg20662988) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.62E+00 | Statistic Test | p-value: 4.37E-04; Z-score: 2.72E+00 | ||
|
Methylation in Case |
3.52E-01 (Median) | Methylation in Control | 2.17E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 10 |
Methylation of SLC5A5 in colon adenocarcinoma | [ 5 ] | |||
|
Location |
Body (cg13805730) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.11E+00 | Statistic Test | p-value: 9.86E-04; Z-score: -1.80E+00 | ||
|
Methylation in Case |
7.20E-01 (Median) | Methylation in Control | 8.02E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 11 |
Methylation of SLC5A5 in colon adenocarcinoma | [ 5 ] | |||
|
Location |
Body (cg11112833) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.04E+00 | Statistic Test | p-value: 2.01E-03; Z-score: -1.11E+00 | ||
|
Methylation in Case |
8.27E-01 (Median) | Methylation in Control | 8.62E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 12 |
Methylation of SLC5A5 in colon adenocarcinoma | [ 5 ] | |||
|
Location |
Body (cg21181547) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.08E+00 | Statistic Test | p-value: 2.11E-03; Z-score: -2.60E+00 | ||
|
Methylation in Case |
7.66E-01 (Median) | Methylation in Control | 8.28E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colorectal cancer |
13 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC5A5 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS1500 (cg10017118) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.11E+00 | Statistic Test | p-value: 6.23E-11; Z-score: -2.78E+00 | ||
|
Methylation in Case |
6.83E-01 (Median) | Methylation in Control | 7.59E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC5A5 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS200 (cg23222039) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.32E+00 | Statistic Test | p-value: 1.73E-07; Z-score: 1.75E+00 | ||
|
Methylation in Case |
1.83E-01 (Median) | Methylation in Control | 1.39E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC5A5 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS200 (cg24065770) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.32E+00 | Statistic Test | p-value: 3.17E-05; Z-score: 8.46E-01 | ||
|
Methylation in Case |
2.16E-02 (Median) | Methylation in Control | 1.63E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC5A5 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS200 (cg21477717) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.12E+00 | Statistic Test | p-value: 8.72E-04; Z-score: 7.23E-01 | ||
|
Methylation in Case |
2.23E-01 (Median) | Methylation in Control | 1.99E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC5A5 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS200 (cg06697339) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.19E+00 | Statistic Test | p-value: 1.13E-03; Z-score: 6.64E-01 | ||
|
Methylation in Case |
3.75E-02 (Median) | Methylation in Control | 3.16E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC5A5 in colorectal cancer | [ 6 ] | |||
|
Location |
1stExon (cg24881657) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.41E+00 | Statistic Test | p-value: 1.11E-07; Z-score: 2.12E+00 | ||
|
Methylation in Case |
1.94E-01 (Median) | Methylation in Control | 1.38E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC5A5 in colorectal cancer | [ 6 ] | |||
|
Location |
Body (cg22179256) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.02E+00 | Statistic Test | p-value: 8.33E-06; Z-score: -1.81E+00 | ||
|
Methylation in Case |
8.77E-01 (Median) | Methylation in Control | 8.98E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC5A5 in colorectal cancer | [ 6 ] | |||
|
Location |
Body (cg21522295) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.34E+00 | Statistic Test | p-value: 4.18E-04; Z-score: 1.20E+00 | ||
|
Methylation in Case |
2.39E-01 (Median) | Methylation in Control | 1.78E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLC5A5 in colorectal cancer | [ 6 ] | |||
|
Location |
Body (cg10660916) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.11E+00 | Statistic Test | p-value: 5.10E-04; Z-score: 7.05E-01 | ||
|
Methylation in Case |
3.30E-01 (Median) | Methylation in Control | 2.96E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 10 |
Methylation of SLC5A5 in colorectal cancer | [ 6 ] | |||
|
Location |
Body (cg15427234) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.13E+00 | Statistic Test | p-value: 5.45E-03; Z-score: -5.79E-01 | ||
|
Methylation in Case |
1.57E-01 (Median) | Methylation in Control | 1.78E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 11 |
Methylation of SLC5A5 in colorectal cancer | [ 6 ] | |||
|
Location |
Body (cg12122403) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.03E+00 | Statistic Test | p-value: 4.87E-02; Z-score: 2.17E-01 | ||
|
Methylation in Case |
1.10E-01 (Median) | Methylation in Control | 1.06E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 12 |
Methylation of SLC5A5 in colorectal cancer | [ 6 ] | |||
|
Location |
3'UTR (cg12562245) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.09E+00 | Statistic Test | p-value: 1.32E-08; Z-score: -3.62E+00 | ||
|
Methylation in Case |
8.21E-01 (Median) | Methylation in Control | 8.97E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Hepatocellular carcinoma |
16 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC5A5 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS1500 (cg27645544) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.64E+00 | Statistic Test | p-value: 5.09E-18; Z-score: -3.45E+00 | ||
|
Methylation in Case |
3.58E-01 (Median) | Methylation in Control | 5.85E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC5A5 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS1500 (cg03640993) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.44E+00 | Statistic Test | p-value: 1.31E-10; Z-score: -3.41E+00 | ||
|
Methylation in Case |
5.03E-01 (Median) | Methylation in Control | 7.22E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC5A5 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS1500 (cg21396646) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.27E+00 | Statistic Test | p-value: 6.22E-10; Z-score: -3.60E+00 | ||
|
Methylation in Case |
5.88E-01 (Median) | Methylation in Control | 7.46E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC5A5 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS1500 (cg17933226) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.21E+00 | Statistic Test | p-value: 6.50E-08; Z-score: -1.61E+00 | ||
|
Methylation in Case |
5.14E-01 (Median) | Methylation in Control | 6.23E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC5A5 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg16701007) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.31E+00 | Statistic Test | p-value: 1.11E-07; Z-score: -2.28E+00 | ||
|
Methylation in Case |
3.32E-01 (Median) | Methylation in Control | 4.35E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC5A5 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg21477717) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.25E+00 | Statistic Test | p-value: 3.78E-06; Z-score: -1.04E+00 | ||
|
Methylation in Case |
1.53E-01 (Median) | Methylation in Control | 1.90E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC5A5 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg01655355) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.17E+00 | Statistic Test | p-value: 1.45E-04; Z-score: -5.55E-01 | ||
|
Methylation in Case |
5.42E-02 (Median) | Methylation in Control | 6.36E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC5A5 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg23222039) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.23E+00 | Statistic Test | p-value: 3.55E-04; Z-score: -9.23E-01 | ||
|
Methylation in Case |
1.21E-01 (Median) | Methylation in Control | 1.49E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLC5A5 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg24065770) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.40E+00 | Statistic Test | p-value: 5.61E-03; Z-score: -5.61E-01 | ||
|
Methylation in Case |
3.08E-02 (Median) | Methylation in Control | 4.31E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 10 |
Methylation of SLC5A5 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg02674588) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.07E+00 | Statistic Test | p-value: 1.14E-02; Z-score: -3.67E-01 | ||
|
Methylation in Case |
8.01E-02 (Median) | Methylation in Control | 8.54E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 11 |
Methylation of SLC5A5 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
1stExon (cg24881657) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.08E+00 | Statistic Test | p-value: 1.57E-03; Z-score: 3.53E-01 | ||
|
Methylation in Case |
1.88E-01 (Median) | Methylation in Control | 1.75E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 12 |
Methylation of SLC5A5 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
Body (cg25996290) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.34E+00 | Statistic Test | p-value: 1.79E-12; Z-score: -5.24E+00 | ||
|
Methylation in Case |
5.73E-01 (Median) | Methylation in Control | 7.66E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 13 |
Methylation of SLC5A5 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
Body (cg25429150) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.32E+00 | Statistic Test | p-value: 1.12E-11; Z-score: -3.55E+00 | ||
|
Methylation in Case |
5.59E-01 (Median) | Methylation in Control | 7.37E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 14 |
Methylation of SLC5A5 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
Body (cg06807030) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.20E+00 | Statistic Test | p-value: 1.62E-11; Z-score: -3.26E+00 | ||
|
Methylation in Case |
6.18E-01 (Median) | Methylation in Control | 7.42E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 15 |
Methylation of SLC5A5 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
Body (cg10359333) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.02E+00 | Statistic Test | p-value: 1.69E-06; Z-score: -1.29E+00 | ||
|
Methylation in Case |
8.75E-01 (Median) | Methylation in Control | 8.95E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 16 |
Methylation of SLC5A5 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
Body (cg11340363) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.07E+00 | Statistic Test | p-value: 5.45E-03; Z-score: 1.59E-01 | ||
|
Methylation in Case |
4.61E-02 (Median) | Methylation in Control | 4.29E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Lung adenocarcinoma |
6 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC5A5 in lung adenocarcinoma | [ 8 ] | |||
|
Location |
TSS1500 (cg17933226) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.06E+00 | Statistic Test | p-value: 4.51E-02; Z-score: 1.02E+00 | ||
|
Methylation in Case |
7.24E-01 (Median) | Methylation in Control | 6.82E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC5A5 in lung adenocarcinoma | [ 8 ] | |||
|
Location |
TSS200 (cg21477717) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.30E+00 | Statistic Test | p-value: 2.33E-02; Z-score: 2.56E+00 | ||
|
Methylation in Case |
2.52E-01 (Median) | Methylation in Control | 1.94E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC5A5 in lung adenocarcinoma | [ 8 ] | |||
|
Location |
TSS200 (cg16701007) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.19E+00 | Statistic Test | p-value: 2.87E-02; Z-score: 1.37E+00 | ||
|
Methylation in Case |
5.46E-01 (Median) | Methylation in Control | 4.61E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC5A5 in lung adenocarcinoma | [ 8 ] | |||
|
Location |
TSS200 (cg23222039) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.45E+00 | Statistic Test | p-value: 3.03E-02; Z-score: 2.33E+00 | ||
|
Methylation in Case |
2.12E-01 (Median) | Methylation in Control | 1.46E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC5A5 in lung adenocarcinoma | [ 8 ] | |||
|
Location |
1stExon (cg24881657) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.86E+00 | Statistic Test | p-value: 4.35E-03; Z-score: 5.05E+00 | ||
|
Methylation in Case |
3.43E-01 (Median) | Methylation in Control | 1.84E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC5A5 in lung adenocarcinoma | [ 8 ] | |||
|
Location |
Body (cg21522295) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.52E+00 | Statistic Test | p-value: 3.80E-02; Z-score: 3.23E+00 | ||
|
Methylation in Case |
3.09E-01 (Median) | Methylation in Control | 2.03E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Prostate cancer |
7 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC5A5 in prostate cancer | [ 9 ] | |||
|
Location |
TSS1500 (cg11493924) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.25E+00 | Statistic Test | p-value: 4.45E-02; Z-score: -1.68E+00 | ||
|
Methylation in Case |
6.77E-02 (Median) | Methylation in Control | 8.47E-02 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC5A5 in prostate cancer | [ 9 ] | |||
|
Location |
TSS1500 (cg25507121) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.43E+00 | Statistic Test | p-value: 4.71E-02; Z-score: -3.13E+00 | ||
|
Methylation in Case |
4.66E-01 (Median) | Methylation in Control | 6.67E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC5A5 in prostate cancer | [ 9 ] | |||
|
Location |
TSS200 (cg03045620) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.20E+00 | Statistic Test | p-value: 2.36E-02; Z-score: -4.34E+00 | ||
|
Methylation in Case |
3.83E-01 (Median) | Methylation in Control | 4.60E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC5A5 in prostate cancer | [ 9 ] | |||
|
Location |
Body (cg01922613) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.41E+00 | Statistic Test | p-value: 8.96E-03; Z-score: 1.20E+01 | ||
|
Methylation in Case |
7.17E-01 (Median) | Methylation in Control | 5.07E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC5A5 in prostate cancer | [ 9 ] | |||
|
Location |
Body (cg00232679) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.40E+00 | Statistic Test | p-value: 1.32E-02; Z-score: 2.72E+00 | ||
|
Methylation in Case |
6.15E-01 (Median) | Methylation in Control | 4.38E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC5A5 in prostate cancer | [ 9 ] | |||
|
Location |
Body (cg23337754) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -2.58E+00 | Statistic Test | p-value: 4.76E-02; Z-score: -1.81E+00 | ||
|
Methylation in Case |
6.94E-02 (Median) | Methylation in Control | 1.79E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
HIV infection |
12 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC5A5 in HIV infection | [ 10 ] | |||
|
Location |
TSS200 (cg21477717) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.33E+00 | Statistic Test | p-value: 8.70E-07; Z-score: 1.64E+00 | ||
|
Methylation in Case |
2.29E-01 (Median) | Methylation in Control | 1.72E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC5A5 in HIV infection | [ 10 ] | |||
|
Location |
TSS200 (cg24065770) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 2.19E+00 | Statistic Test | p-value: 2.50E-06; Z-score: 2.54E+00 | ||
|
Methylation in Case |
7.46E-02 (Median) | Methylation in Control | 3.40E-02 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC5A5 in HIV infection | [ 10 ] | |||
|
Location |
TSS200 (cg23222039) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.56E+00 | Statistic Test | p-value: 1.20E-05; Z-score: 2.27E+00 | ||
|
Methylation in Case |
1.79E-01 (Median) | Methylation in Control | 1.15E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC5A5 in HIV infection | [ 10 ] | |||
|
Location |
TSS200 (cg06697339) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.79E+00 | Statistic Test | p-value: 2.85E-05; Z-score: 2.20E+00 | ||
|
Methylation in Case |
6.86E-02 (Median) | Methylation in Control | 3.84E-02 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC5A5 in HIV infection | [ 10 ] | |||
|
Location |
TSS200 (cg01655355) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.46E+00 | Statistic Test | p-value: 3.52E-04; Z-score: 1.65E+00 | ||
|
Methylation in Case |
7.71E-02 (Median) | Methylation in Control | 5.28E-02 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC5A5 in HIV infection | [ 10 ] | |||
|
Location |
TSS200 (cg02674588) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.22E+00 | Statistic Test | p-value: 5.23E-03; Z-score: 1.17E+00 | ||
|
Methylation in Case |
9.53E-02 (Median) | Methylation in Control | 7.79E-02 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC5A5 in HIV infection | [ 10 ] | |||
|
Location |
1stExon (cg24881657) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.72E+00 | Statistic Test | p-value: 2.14E-06; Z-score: 2.70E+00 | ||
|
Methylation in Case |
2.09E-01 (Median) | Methylation in Control | 1.22E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC5A5 in HIV infection | [ 10 ] | |||
|
Location |
Body (cg10660916) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.29E+00 | Statistic Test | p-value: 4.06E-08; Z-score: 1.87E+00 | ||
|
Methylation in Case |
3.39E-01 (Median) | Methylation in Control | 2.63E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLC5A5 in HIV infection | [ 10 ] | |||
|
Location |
Body (cg15427234) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.25E+00 | Statistic Test | p-value: 3.85E-04; Z-score: 1.36E+00 | ||
|
Methylation in Case |
1.27E-01 (Median) | Methylation in Control | 1.01E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 10 |
Methylation of SLC5A5 in HIV infection | [ 10 ] | |||
|
Location |
Body (cg21522295) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.14E+00 | Statistic Test | p-value: 9.22E-03; Z-score: 8.64E-01 | ||
|
Methylation in Case |
1.09E-01 (Median) | Methylation in Control | 9.54E-02 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 11 |
Methylation of SLC5A5 in HIV infection | [ 10 ] | |||
|
Location |
Body (cg22179256) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.01E+00 | Statistic Test | p-value: 2.07E-02; Z-score: -4.68E-01 | ||
|
Methylation in Case |
8.52E-01 (Median) | Methylation in Control | 8.63E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 12 |
Methylation of SLC5A5 in HIV infection | [ 10 ] | |||
|
Location |
Body (cg11340363) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.41E+00 | Statistic Test | p-value: 2.44E-02; Z-score: 6.05E-01 | ||
|
Methylation in Case |
4.02E-02 (Median) | Methylation in Control | 2.84E-02 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Panic disorder |
4 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC5A5 in panic disorder | [ 11 ] | |||
|
Location |
TSS200 (cg21477717) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -9.34E-01 | Statistic Test | p-value: 1.85E-04; Z-score: -4.98E-01 | ||
|
Methylation in Case |
-3.07E+00 (Median) | Methylation in Control | -2.87E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder [ ICD-11: 6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC5A5 in panic disorder | [ 11 ] | |||
|
Location |
TSS200 (cg24065770) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -9.64E-01 | Statistic Test | p-value: 4.45E-02; Z-score: -4.21E-01 | ||
|
Methylation in Case |
-5.88E+00 (Median) | Methylation in Control | -5.67E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder [ ICD-11: 6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC5A5 in panic disorder | [ 11 ] | |||
|
Location |
1stExon (cg24881657) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -9.41E-01 | Statistic Test | p-value: 2.23E-02; Z-score: -4.40E-01 | ||
|
Methylation in Case |
-3.64E+00 (Median) | Methylation in Control | -3.42E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder [ ICD-11: 6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC5A5 in panic disorder | [ 11 ] | |||
|
Location |
Body (cg10660916) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -9.12E-01 | Statistic Test | p-value: 1.10E-02; Z-score: -3.15E-01 | ||
|
Methylation in Case |
-1.94E+00 (Median) | Methylation in Control | -1.77E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder [ ICD-11: 6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Papillary thyroid cancer |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC5A5 in papillary thyroid cancer | [ 12 ] | |||
|
Location |
TSS200 (cg23222039) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.36E+00 | Statistic Test | p-value: 5.23E-04; Z-score: -9.79E-01 | ||
|
Methylation in Case |
1.02E-01 (Median) | Methylation in Control | 1.39E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC5A5 in papillary thyroid cancer | [ 12 ] | |||
|
Location |
Body (cg22179256) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.01E+00 | Statistic Test | p-value: 3.63E-03; Z-score: -5.41E-01 | ||
|
Methylation in Case |
8.67E-01 (Median) | Methylation in Control | 8.78E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC5A5 in papillary thyroid cancer | [ 12 ] | |||
|
Location |
Body (cg21522295) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.23E+00 | Statistic Test | p-value: 4.21E-03; Z-score: -7.73E-01 | ||
|
Methylation in Case |
1.34E-01 (Median) | Methylation in Control | 1.65E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Systemic lupus erythematosus |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC5A5 in systemic lupus erythematosus | [ 13 ] | |||
|
Location |
TSS200 (cg06697339) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.06E+00 | Statistic Test | p-value: 1.28E-02; Z-score: 8.76E-02 | ||
|
Methylation in Case |
4.95E-02 (Median) | Methylation in Control | 4.69E-02 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus [ ICD-11: 4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC5A5 in systemic lupus erythematosus | [ 13 ] | |||
|
Location |
Body (cg21522295) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.06E+00 | Statistic Test | p-value: 1.09E-02; Z-score: -2.56E-01 | ||
|
Methylation in Case |
1.12E-01 (Median) | Methylation in Control | 1.18E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus [ ICD-11: 4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC5A5 in systemic lupus erythematosus | [ 13 ] | |||
|
Location |
Body (cg10660916) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.05E+00 | Statistic Test | p-value: 2.58E-02; Z-score: 2.67E-01 | ||
|
Methylation in Case |
3.01E-01 (Median) | Methylation in Control | 2.87E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus [ ICD-11: 4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Atypical teratoid rhabdoid tumor |
6 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC5A5 in atypical teratoid rhabdoid tumor | [ 14 ] | |||
|
Location |
1stExon (cg24881657) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.27E+00 | Statistic Test | p-value: 2.03E-16; Z-score: -2.87E+00 | ||
|
Methylation in Case |
6.38E-01 (Median) | Methylation in Control | 8.13E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC5A5 in atypical teratoid rhabdoid tumor | [ 14 ] | |||
|
Location |
Body (cg10359333) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.05E+00 | Statistic Test | p-value: 1.44E-02; Z-score: -5.16E-01 | ||
|
Methylation in Case |
8.28E-01 (Median) | Methylation in Control | 8.69E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC5A5 in atypical teratoid rhabdoid tumor | [ 14 ] | |||
|
Location |
Body (cg10660916) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.27E+00 | Statistic Test | p-value: 1.70E-02; Z-score: 9.07E-01 | ||
|
Methylation in Case |
7.47E-01 (Median) | Methylation in Control | 5.89E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC5A5 in atypical teratoid rhabdoid tumor | [ 14 ] | |||
|
Location |
Body (cg11340363) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.01E+00 | Statistic Test | p-value: 2.14E-02; Z-score: -2.41E-01 | ||
|
Methylation in Case |
8.70E-01 (Median) | Methylation in Control | 8.81E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC5A5 in atypical teratoid rhabdoid tumor | [ 14 ] | |||
|
Location |
Body (cg12122403) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.43E+00 | Statistic Test | p-value: 2.67E-02; Z-score: -6.07E-01 | ||
|
Methylation in Case |
1.12E-01 (Median) | Methylation in Control | 1.60E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC5A5 in atypical teratoid rhabdoid tumor | [ 14 ] | |||
|
Location |
3'UTR (cg12562245) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.15E+00 | Statistic Test | p-value: 2.36E-11; Z-score: -2.21E+00 | ||
|
Methylation in Case |
7.78E-01 (Median) | Methylation in Control | 8.92E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Depression |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC5A5 in depression | [ 15 ] | |||
|
Location |
Body (cg10660916) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.08E+00 | Statistic Test | p-value: 4.77E-02; Z-score: 5.86E-01 | ||
|
Methylation in Case |
2.63E-01 (Median) | Methylation in Control | 2.44E-01 (Median) | ||
|
Studied Phenotype |
Depression [ ICD-11: 6A8Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Esthesioneuroblastoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypermethylation of SLC5A5 in esthesioneuroblastoma than that in healthy individual | ||||
Studied Phenotype |
Esthesioneuroblastoma [ICD-11:2D50.1] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 1.96E-08; Fold-change: 0.365915694; Z-score: 3.108466779 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
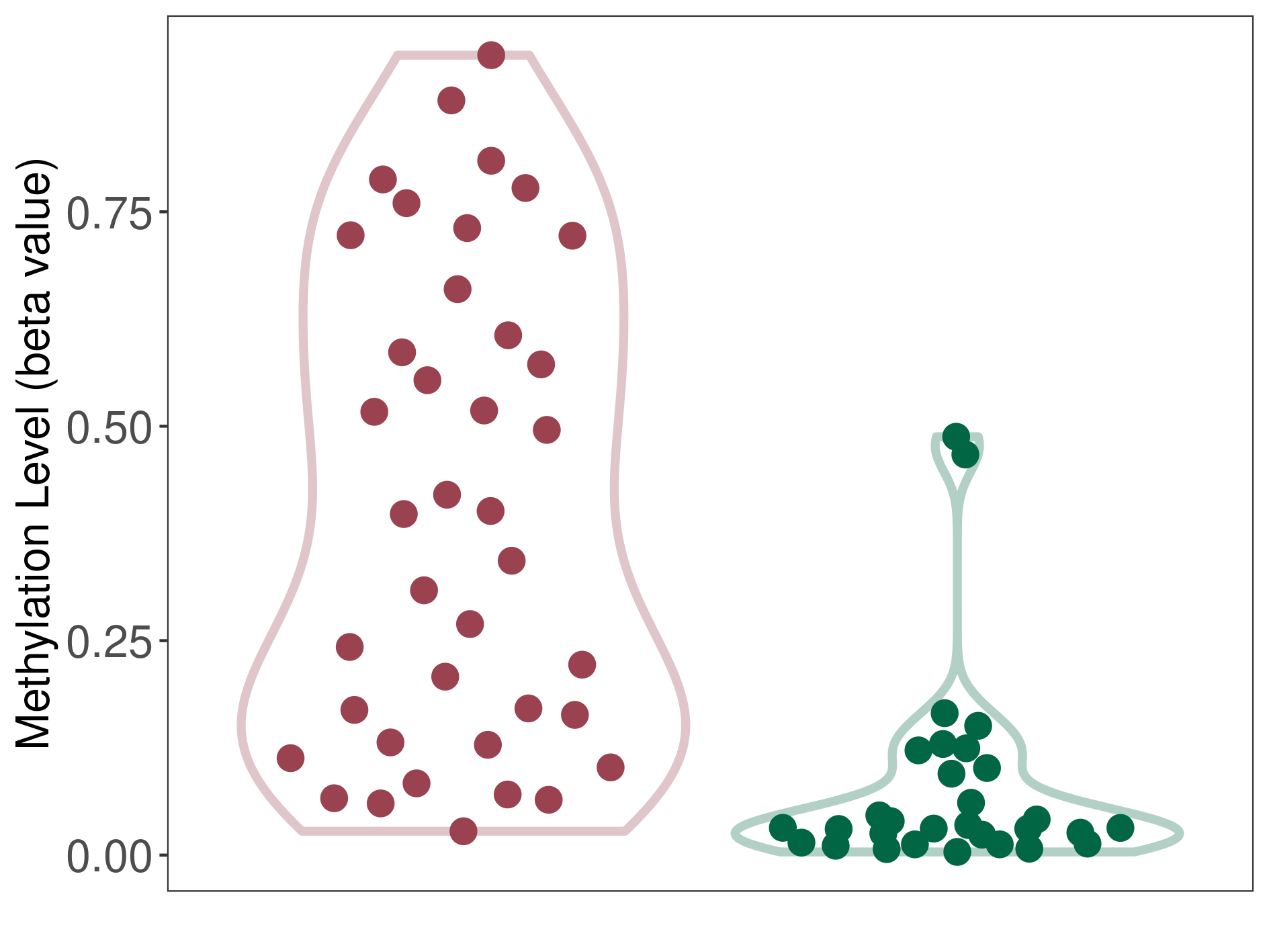
|
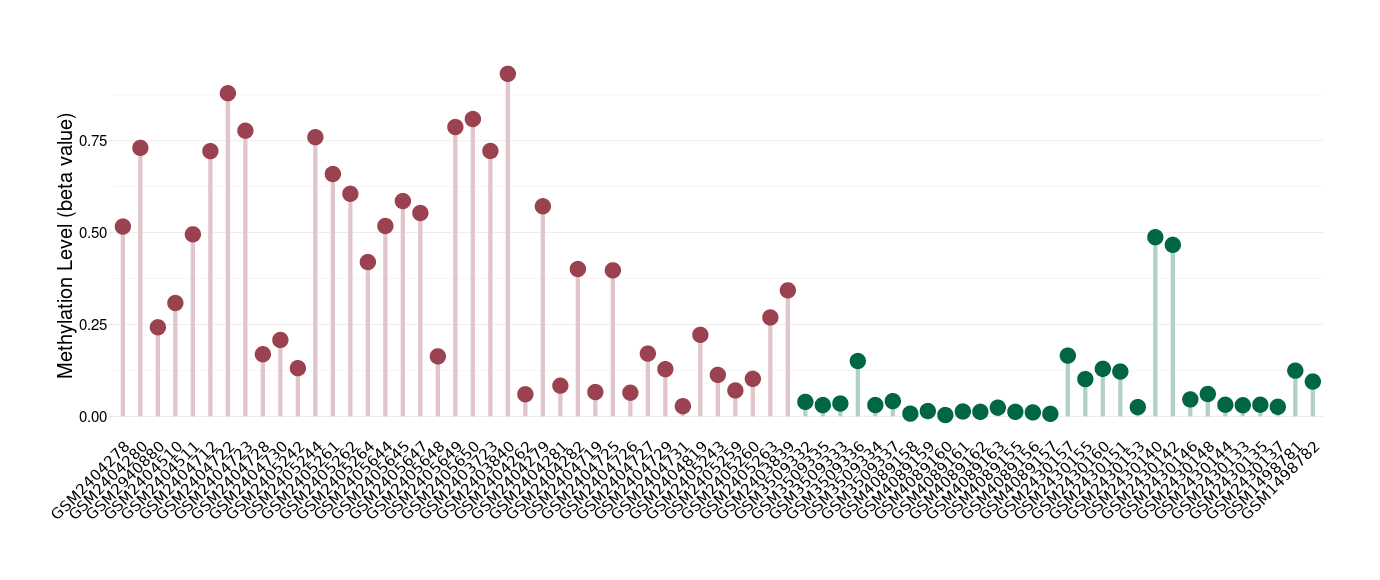 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Lymphoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypermethylation of SLC5A5 in lymphoma than that in healthy individual | ||||
Studied Phenotype |
Lymphoma [ICD-11:2B30] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 2.16E-09; Fold-change: 0.310791842; Z-score: 3.774794276 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
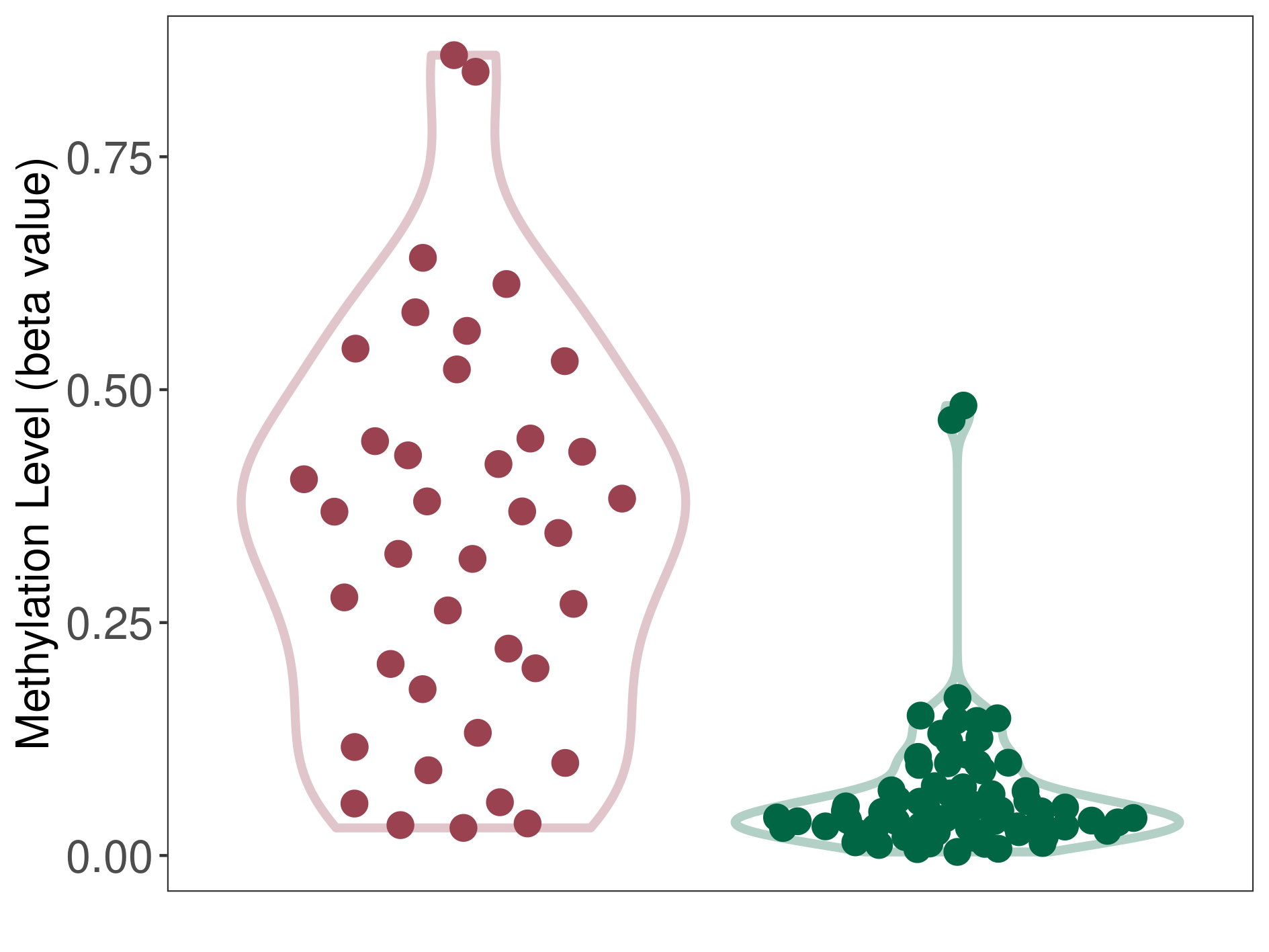
|
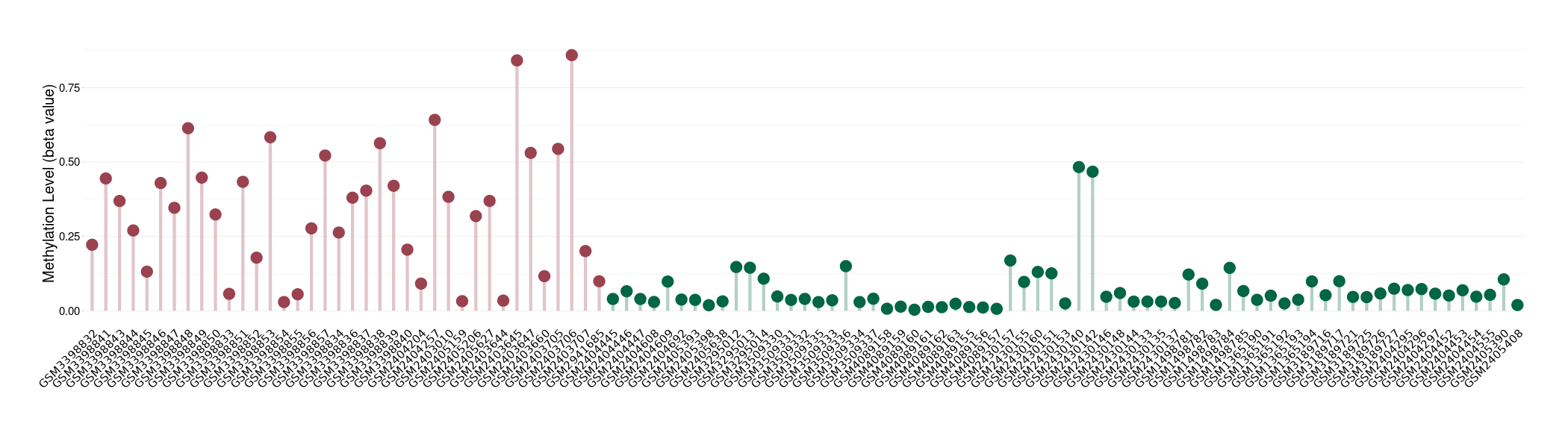 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Histone acetylation |
|||||
|
Breast cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Hypoacetylation of SLC5A5 in breast cancer (compare with SAHA treatment counterpart cells) | [ 16 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes |
Down regulation of SLC5A5 | Experiment Method | Western Blot | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
Levels of acetylated K9 and K14 were increased in high-responsive cells (MDA468) and reduced in low-responsive cells (MDA157). It is possible to hypothesize that decrease of H3 histone acetylation in low-responsive cells is not permissive for induction of high levels of NIS mRNA upon SAHA treatment. | ||||
|
Thyroid cancer |
7 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Hypoacetylation of SLC5A5 in thyroid cancer (compare with counterpart healthy thyroid cells) | [ 17 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes |
Down regulation of SLC5A5 | Experiment Method | Western Blot | ||
|
Studied Phenotype |
Thyroid cancer [ ICD-11: 2D10] | ||||
|
Experimental Material |
Multiple cell lines of human and rat | ||||
|
Additional Notes |
Histone deacetylation occurred on all these lysine residues of histones at the SLC5A5 promoter upon the activation of MAPK by BRAF V600E. | ||||
|
Epigenetic Phenomenon 2 |
Hypoacetylation of SLC5A5 in thyroid cancer (compare with counterpart healthy thyroid cells) | [ 17 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes |
Down regulation of SLC5A5 | Experiment Method | Western Blot | ||
|
Studied Phenotype |
Thyroid cancer [ ICD-11: 2D10] | ||||
|
Experimental Material |
Multiple cell lines of human and rat | ||||
|
Additional Notes |
Histone deacetylation occurred on all these lysine residues of histones at the SLC5A5 promoter upon the activation of MAPK by BRAF V600E. | ||||
|
Epigenetic Phenomenon 3 |
Hypoacetylation of SLC5A5 in thyroid cancer (compare with counterpart healthy thyroid cells) | [ 17 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes |
Down regulation of SLC5A5 | Experiment Method | Western Blot | ||
|
Studied Phenotype |
Thyroid cancer [ ICD-11: 2D10] | ||||
|
Experimental Material |
Multiple cell lines of human and rat | ||||
|
Additional Notes |
Histone deacetylation occurred on all these lysine residues of histones at the SLC5A5 promoter upon the activation of MAPK by BRAF V600E. | ||||
|
Epigenetic Phenomenon 4 |
Hypoacetylation of SLC5A5 in thyroid cancer (compare with counterpart healthy thyroid cells) | [ 17 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes |
Down regulation of SLC5A5 | Experiment Method | Western Blot | ||
|
Studied Phenotype |
Thyroid cancer [ ICD-11: 2D10] | ||||
|
Experimental Material |
Multiple cell lines of human and rat | ||||
|
Additional Notes |
Histone deacetylation occurred on all these lysine residues of histones at the SLC5A5 promoter upon the activation of MAPK by BRAF V600E. | ||||
|
Epigenetic Phenomenon 5 |
Hypoacetylation of SLC5A5 in thyroid cancer (compare with counterpart healthy thyroid cells) | [ 18 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes |
Down regulation of SLC5A5 | Experiment Method | Western Blot | ||
|
Studied Phenotype |
Thyroid cancer [ ICD-11: 2D10] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
The underlying mechanism of HDACi-induced upregulation of SLC5A5 gene expression via improving the acetylation status of histone at the promoter was elucidated directly by ChIP assay | ||||
|
Epigenetic Phenomenon 6 |
Hypoacetylation of SLC5A5 in thyroid cancer (compare with counterpart healthy thyroid cells) | [ 18 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes |
Down regulation of SLC5A5 | Experiment Method | Western Blot | ||
|
Studied Phenotype |
Thyroid cancer [ ICD-11: 2D10] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
The underlying mechanism of HDACi-induced upregulation of SLC5A5 gene expression via improving the acetylation status of histone at the promoter was elucidated directly by ChIP assay | ||||
|
Epigenetic Phenomenon 7 |
Hypoacetylation of SLC5A5 in thyroid cancer (compare with counterpart healthy thyroid cells) | [ 18 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes |
Down regulation of SLC5A5 | Experiment Method | Western Blot | ||
|
Studied Phenotype |
Thyroid cancer [ ICD-11: 2D10] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
The underlying mechanism of HDACi-induced upregulation of SLC5A5 gene expression via improving the acetylation status of histone at the promoter was elucidated directly by ChIP assay | ||||
|
microRNA |
|||||
|
Unclear Phenotype |
44 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
miR-10a directly targets SLC5A5 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-10a | miRNA Mature ID | miR-10a-5p | ||
|
miRNA Sequence |
UACCCUGUAGAUCCGAAUUUGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 2 |
miR-10b directly targets SLC5A5 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-10b | miRNA Mature ID | miR-10b-5p | ||
|
miRNA Sequence |
UACCCUGUAGAACCGAAUUUGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 3 |
miR-1228 directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-1228 | miRNA Mature ID | miR-1228-3p | ||
|
miRNA Sequence |
UCACACCUGCCUCGCCCCCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 4 |
miR-149 directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-149 | miRNA Mature ID | miR-149-3p | ||
|
miRNA Sequence |
AGGGAGGGACGGGGGCUGUGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 5 |
miR-1827 directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-1827 | miRNA Mature ID | miR-1827 | ||
|
miRNA Sequence |
UGAGGCAGUAGAUUGAAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 6 |
miR-1910 directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-1910 | miRNA Mature ID | miR-1910-3p | ||
|
miRNA Sequence |
GAGGCAGAAGCAGGAUGACA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 7 |
miR-2114 directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-2114 | miRNA Mature ID | miR-2114-3p | ||
|
miRNA Sequence |
CGAGCCUCAAGCAAGGGACUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 8 |
miR-2467 directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-2467 | miRNA Mature ID | miR-2467-3p | ||
|
miRNA Sequence |
AGCAGAGGCAGAGAGGCUCAGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 9 |
miR-30b directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-30b | miRNA Mature ID | miR-30b-3p | ||
|
miRNA Sequence |
CUGGGAGGUGGAUGUUUACUUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 10 |
miR-3192 directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3192 | miRNA Mature ID | miR-3192-5p | ||
|
miRNA Sequence |
UCUGGGAGGUUGUAGCAGUGGAA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 11 |
miR-339 directly targets SLC5A5 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-339 | miRNA Mature ID | miR-339-5p | ||
|
miRNA Sequence |
UCCCUGUCCUCCAGGAGCUCACG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 12 |
miR-34b directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-34b | miRNA Mature ID | miR-34b-3p | ||
|
miRNA Sequence |
CAAUCACUAACUCCACUGCCAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 13 |
miR-3612 directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3612 | miRNA Mature ID | miR-3612 | ||
|
miRNA Sequence |
AGGAGGCAUCUUGAGAAAUGGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 14 |
miR-3614 directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3614 | miRNA Mature ID | miR-3614-5p | ||
|
miRNA Sequence |
CCACUUGGAUCUGAAGGCUGCCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 15 |
miR-3689c directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3689c | miRNA Mature ID | miR-3689c | ||
|
miRNA Sequence |
CUGGGAGGUGUGAUAUUGUGGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 16 |
miR-377 directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-377 | miRNA Mature ID | miR-377-5p | ||
|
miRNA Sequence |
AGAGGUUGCCCUUGGUGAAUUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 17 |
miR-4257 directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4257 | miRNA Mature ID | miR-4257 | ||
|
miRNA Sequence |
CCAGAGGUGGGGACUGAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 18 |
miR-4421 directly targets SLC5A5 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4421 | miRNA Mature ID | miR-4421 | ||
|
miRNA Sequence |
ACCUGUCUGUGGAAAGGAGCUA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 19 |
miR-4537 directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4537 | miRNA Mature ID | miR-4537 | ||
|
miRNA Sequence |
UGAGCCGAGCUGAGCUUAGCUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 20 |
miR-455 directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-455 | miRNA Mature ID | miR-455-3p | ||
|
miRNA Sequence |
GCAGUCCAUGGGCAUAUACAC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 21 |
miR-4684 directly targets SLC5A5 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4684 | miRNA Mature ID | miR-4684-5p | ||
|
miRNA Sequence |
CUCUCUACUGACUUGCAACAUA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 22 |
miR-4715 directly targets SLC5A5 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4715 | miRNA Mature ID | miR-4715-5p | ||
|
miRNA Sequence |
AAGUUGGCUGCAGUUAAGGUGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 23 |
miR-4728 directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4728 | miRNA Mature ID | miR-4728-5p | ||
|
miRNA Sequence |
UGGGAGGGGAGAGGCAGCAAGCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 24 |
miR-548c directly targets SLC5A5 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-548c | miRNA Mature ID | miR-548c-3p | ||
|
miRNA Sequence |
CAAAAAUCUCAAUUACUUUUGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 25 |
miR-5699 directly targets SLC5A5 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-5699 | miRNA Mature ID | miR-5699-3p | ||
|
miRNA Sequence |
UCCUGUCUUUCCUUGUUGGAGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 26 |
miR-593 directly targets SLC5A5 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-593 | miRNA Mature ID | miR-593-3p | ||
|
miRNA Sequence |
UGUCUCUGCUGGGGUUUCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 27 |
miR-6086 directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6086 | miRNA Mature ID | miR-6086 | ||
|
miRNA Sequence |
GGAGGUUGGGAAGGGCAGAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 28 |
miR-647 directly targets SLC5A5 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-647 | miRNA Mature ID | miR-647 | ||
|
miRNA Sequence |
GUGGCUGCACUCACUUCCUUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 29 |
miR-6499 directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6499 | miRNA Mature ID | miR-6499-3p | ||
|
miRNA Sequence |
AGCAGUGUUUGUUUUGCCCACA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 30 |
miR-650 directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-650 | miRNA Mature ID | miR-650 | ||
|
miRNA Sequence |
AGGAGGCAGCGCUCUCAGGAC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 31 |
miR-6500 directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6500 | miRNA Mature ID | miR-6500-3p | ||
|
miRNA Sequence |
ACACUUGUUGGGAUGACCUGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 32 |
miR-6516 directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6516 | miRNA Mature ID | miR-6516-5p | ||
|
miRNA Sequence |
UUUGCAGUAACAGGUGUGAGCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 33 |
miR-661 directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-661 | miRNA Mature ID | miR-661 | ||
|
miRNA Sequence |
UGCCUGGGUCUCUGGCCUGCGCGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 34 |
miR-6732 directly targets SLC5A5 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6732 | miRNA Mature ID | miR-6732-3p | ||
|
miRNA Sequence |
UAACCCUGUCCUCUCCCUCCCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 35 |
miR-6779 directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6779 | miRNA Mature ID | miR-6779-5p | ||
|
miRNA Sequence |
CUGGGAGGGGCUGGGUUUGGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 36 |
miR-6785 directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6785 | miRNA Mature ID | miR-6785-5p | ||
|
miRNA Sequence |
UGGGAGGGCGUGGAUGAUGGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 37 |
miR-6799 directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6799 | miRNA Mature ID | miR-6799-5p | ||
|
miRNA Sequence |
GGGGAGGUGUGCAGGGCUGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 38 |
miR-6807 directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6807 | miRNA Mature ID | miR-6807-5p | ||
|
miRNA Sequence |
GUGAGCCAGUGGAAUGGAGAGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 39 |
miR-6818 directly targets SLC5A5 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6818 | miRNA Mature ID | miR-6818-3p | ||
|
miRNA Sequence |
UUGUCUCUUGUUCCUCACACAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 40 |
miR-6823 directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6823 | miRNA Mature ID | miR-6823-3p | ||
|
miRNA Sequence |
UGAGCCUCUCCUUCCCUCCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 41 |
miR-6842 directly targets SLC5A5 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6842 | miRNA Mature ID | miR-6842-3p | ||
|
miRNA Sequence |
UUGGCUGGUCUCUGCUCCGCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 42 |
miR-6883 directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6883 | miRNA Mature ID | miR-6883-5p | ||
|
miRNA Sequence |
AGGGAGGGUGUGGUAUGGAUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 43 |
miR-6895 directly targets SLC5A5 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6895 | miRNA Mature ID | miR-6895-3p | ||
|
miRNA Sequence |
UGUCUCUCGCCCUUGGCCUUAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 44 |
miR-7106 directly targets SLC5A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-7106 | miRNA Mature ID | miR-7106-5p | ||
|
miRNA Sequence |
UGGGAGGAGGGGAUCUUGGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.
