Detail Information of Epigenetic Regulations
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0456 Transporter Info | ||||
| Gene Name | SLC6A4 | ||||
| Transporter Name | Sodium-dependent serotonin transporter | ||||
| Gene ID | |||||
| UniProt ID | |||||
| Epigenetic Regulations of This DT (EGR) | |||||
|---|---|---|---|---|---|
|
Methylation |
|||||
|
Gestational diabetes |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Hypomethylation of SLC6A4 in gestational diabetes | [ 1 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Bisulfite sequencing | ||
|
Related Molecular Changes |
Up regulation of SLC6A4 | Experiment Method | RT-qPCR | ||
|
Studied Phenotype |
Gestational diabetes [ ICD-11: JA63.2] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Additional Notes |
Placental SLC6A4 mRNA levels were inversely correlated with average DNA methylation (p = 0.010) while no statistically significant association was found with the SLC6A4 genotypes (p>0.05). | ||||
|
Adiposity |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Hypomethylation of SLC6A4 in adiposity | [ 2 ] | |||
|
Location |
CpG5 | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Bisulfite pyrosequencing | ||
|
Related Molecular Changes |
Down regulation of SLC6A4 | Experiment Method | RT-qPCR | ||
|
Studied Phenotype |
Adiposity [ ICD-11: 5B80-5B81] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Additional Notes |
Methylation of both SLC6A4 CpG5 and expression of SLC6A4 was lower in obese compared with lean adults. | ||||
|
Atypical teratoid rhabdoid tumor |
6 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC6A4 in atypical teratoid rhabdoid tumor | [ 3 ] | |||
|
Location |
5'UTR (cg01330016) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.34E+00 | Statistic Test | p-value: 9.61E-09; Z-score: 1.46E+00 | ||
|
Methylation in Case |
6.38E-01 (Median) | Methylation in Control | 4.76E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC6A4 in atypical teratoid rhabdoid tumor | [ 3 ] | |||
|
Location |
5'UTR (cg03363743) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.12E+00 | Statistic Test | p-value: 1.73E-08; Z-score: -1.68E+00 | ||
|
Methylation in Case |
7.97E-01 (Median) | Methylation in Control | 8.97E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC6A4 in atypical teratoid rhabdoid tumor | [ 3 ] | |||
|
Location |
5'UTR (cg05951817) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.06E+00 | Statistic Test | p-value: 5.13E-08; Z-score: -1.22E+00 | ||
|
Methylation in Case |
8.03E-01 (Median) | Methylation in Control | 8.54E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC6A4 in atypical teratoid rhabdoid tumor | [ 3 ] | |||
|
Location |
5'UTR (cg22584138) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.34E+00 | Statistic Test | p-value: 6.27E-06; Z-score: -1.30E+00 | ||
|
Methylation in Case |
4.71E-01 (Median) | Methylation in Control | 6.32E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC6A4 in atypical teratoid rhabdoid tumor | [ 3 ] | |||
|
Location |
5'UTR (cg26126367) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.06E+00 | Statistic Test | p-value: 1.14E-05; Z-score: -6.64E-01 | ||
|
Methylation in Case |
7.49E-01 (Median) | Methylation in Control | 7.91E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC6A4 in atypical teratoid rhabdoid tumor | [ 3 ] | |||
|
Location |
3'UTR (cg20592995) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.28E+00 | Statistic Test | p-value: 4.21E-10; Z-score: 1.52E+00 | ||
|
Methylation in Case |
7.96E-01 (Median) | Methylation in Control | 6.22E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Bladder cancer |
7 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC6A4 in bladder cancer | [ 4 ] | |||
|
Location |
5'UTR (cg03363743) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.90E+00 | Statistic Test | p-value: 2.18E-06; Z-score: 7.26E+00 | ||
|
Methylation in Case |
4.02E-01 (Median) | Methylation in Control | 2.12E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC6A4 in bladder cancer | [ 4 ] | |||
|
Location |
5'UTR (cg05951817) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.38E+00 | Statistic Test | p-value: 1.78E-02; Z-score: -2.44E+00 | ||
|
Methylation in Case |
2.97E-01 (Median) | Methylation in Control | 4.09E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC6A4 in bladder cancer | [ 4 ] | |||
|
Location |
5'UTR (cg22584138) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.51E+00 | Statistic Test | p-value: 2.42E-02; Z-score: -2.42E+00 | ||
|
Methylation in Case |
2.19E-01 (Median) | Methylation in Control | 3.30E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC6A4 in bladder cancer | [ 4 ] | |||
|
Location |
5'UTR (cg01330016) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.07E+00 | Statistic Test | p-value: 3.84E-02; Z-score: 3.09E+00 | ||
|
Methylation in Case |
7.34E-01 (Median) | Methylation in Control | 6.83E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC6A4 in bladder cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg18584905) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.17E+00 | Statistic Test | p-value: 1.05E-03; Z-score: -3.75E+00 | ||
|
Methylation in Case |
1.00E-01 (Median) | Methylation in Control | 1.17E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC6A4 in bladder cancer | [ 4 ] | |||
|
Location |
TSS200 (cg25725890) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.36E+00 | Statistic Test | p-value: 8.32E-03; Z-score: -2.39E+00 | ||
|
Methylation in Case |
6.34E-02 (Median) | Methylation in Control | 8.65E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC6A4 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg24984698) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.51E+00 | Statistic Test | p-value: 6.03E-10; Z-score: -1.14E+01 | ||
|
Methylation in Case |
4.92E-01 (Median) | Methylation in Control | 7.44E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Breast cancer |
8 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC6A4 in breast cancer | [ 5 ] | |||
|
Location |
5'UTR (cg03363743) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.50E+00 | Statistic Test | p-value: 3.88E-14; Z-score: 1.99E+00 | ||
|
Methylation in Case |
4.04E-01 (Median) | Methylation in Control | 2.69E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC6A4 in breast cancer | [ 5 ] | |||
|
Location |
5'UTR (cg26126367) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.06E+00 | Statistic Test | p-value: 5.81E-07; Z-score: -1.64E+00 | ||
|
Methylation in Case |
7.75E-01 (Median) | Methylation in Control | 8.19E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC6A4 in breast cancer | [ 5 ] | |||
|
Location |
5'UTR (cg05951817) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.07E+00 | Statistic Test | p-value: 1.48E-02; Z-score: -4.58E-01 | ||
|
Methylation in Case |
6.46E-01 (Median) | Methylation in Control | 6.90E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC6A4 in breast cancer | [ 5 ] | |||
|
Location |
TSS1500 (cg18584905) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.39E+00 | Statistic Test | p-value: 3.04E-09; Z-score: 2.45E+00 | ||
|
Methylation in Case |
1.48E-01 (Median) | Methylation in Control | 1.06E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC6A4 in breast cancer | [ 5 ] | |||
|
Location |
TSS1500 (cg06841846) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.43E+00 | Statistic Test | p-value: 4.34E-04; Z-score: 8.11E-01 | ||
|
Methylation in Case |
7.30E-02 (Median) | Methylation in Control | 5.09E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC6A4 in breast cancer | [ 5 ] | |||
|
Location |
TSS1500 (cg12074493) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.16E+00 | Statistic Test | p-value: 1.21E-03; Z-score: 3.52E-01 | ||
|
Methylation in Case |
5.68E-02 (Median) | Methylation in Control | 4.88E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC6A4 in breast cancer | [ 5 ] | |||
|
Location |
TSS200 (cg26741280) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.03E+00 | Statistic Test | p-value: 4.21E-02; Z-score: 2.25E-01 | ||
|
Methylation in Case |
1.77E-01 (Median) | Methylation in Control | 1.72E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC6A4 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg24984698) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.08E+00 | Statistic Test | p-value: 2.59E-06; Z-score: -1.29E+00 | ||
|
Methylation in Case |
6.55E-01 (Median) | Methylation in Control | 7.06E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colorectal cancer |
12 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC6A4 in colorectal cancer | [ 6 ] | |||
|
Location |
5'UTR (cg03363743) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.48E+00 | Statistic Test | p-value: 1.46E-09; Z-score: 2.68E+00 | ||
|
Methylation in Case |
7.07E-01 (Median) | Methylation in Control | 4.77E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC6A4 in colorectal cancer | [ 6 ] | |||
|
Location |
5'UTR (cg22584138) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.40E+00 | Statistic Test | p-value: 1.11E-07; Z-score: 1.97E+00 | ||
|
Methylation in Case |
6.86E-01 (Median) | Methylation in Control | 4.92E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC6A4 in colorectal cancer | [ 6 ] | |||
|
Location |
5'UTR (cg05951817) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.34E+00 | Statistic Test | p-value: 1.86E-07; Z-score: 2.19E+00 | ||
|
Methylation in Case |
7.54E-01 (Median) | Methylation in Control | 5.64E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC6A4 in colorectal cancer | [ 6 ] | |||
|
Location |
5'UTR (cg01330016) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.11E+00 | Statistic Test | p-value: 1.82E-06; Z-score: -1.18E+00 | ||
|
Methylation in Case |
7.68E-01 (Median) | Methylation in Control | 8.56E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC6A4 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS1500 (cg18584905) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.15E+00 | Statistic Test | p-value: 7.17E-05; Z-score: 9.10E-01 | ||
|
Methylation in Case |
2.15E-01 (Median) | Methylation in Control | 1.87E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC6A4 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS1500 (cg06841846) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.01E+00 | Statistic Test | p-value: 1.65E-02; Z-score: 5.34E-02 | ||
|
Methylation in Case |
5.29E-02 (Median) | Methylation in Control | 5.22E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC6A4 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS200 (cg26741280) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.11E+00 | Statistic Test | p-value: 1.89E-05; Z-score: 1.04E+00 | ||
|
Methylation in Case |
1.73E-01 (Median) | Methylation in Control | 1.56E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC6A4 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS200 (cg27569822) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.19E+00 | Statistic Test | p-value: 1.58E-04; Z-score: 1.03E+00 | ||
|
Methylation in Case |
4.38E-02 (Median) | Methylation in Control | 3.67E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLC6A4 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS200 (cg10901968) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.08E+00 | Statistic Test | p-value: 5.13E-04; Z-score: 3.43E-01 | ||
|
Methylation in Case |
3.68E-02 (Median) | Methylation in Control | 3.41E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 10 |
Methylation of SLC6A4 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS200 (cg25725890) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.05E+00 | Statistic Test | p-value: 8.62E-04; Z-score: 2.68E-01 | ||
|
Methylation in Case |
1.07E-01 (Median) | Methylation in Control | 1.02E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 11 |
Methylation of SLC6A4 in colorectal cancer | [ 6 ] | |||
|
Location |
Body (cg24984698) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.06E+00 | Statistic Test | p-value: 1.97E-09; Z-score: -3.02E+00 | ||
|
Methylation in Case |
8.15E-01 (Median) | Methylation in Control | 8.66E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Hepatocellular carcinoma |
17 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC6A4 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
5'UTR (cg26126367) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.12E+00 | Statistic Test | p-value: 6.07E-09; Z-score: -4.26E+00 | ||
|
Methylation in Case |
7.21E-01 (Median) | Methylation in Control | 8.08E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC6A4 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
5'UTR (cg01330016) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.06E+00 | Statistic Test | p-value: 2.40E-07; Z-score: -2.02E+00 | ||
|
Methylation in Case |
7.65E-01 (Median) | Methylation in Control | 8.10E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC6A4 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
5'UTR (cg05951817) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.06E+00 | Statistic Test | p-value: 1.45E-02; Z-score: -4.79E-01 | ||
|
Methylation in Case |
6.66E-01 (Median) | Methylation in Control | 7.03E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC6A4 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS1500 (cg23331484) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.49E+00 | Statistic Test | p-value: 1.59E-13; Z-score: 3.59E+00 | ||
|
Methylation in Case |
6.35E-01 (Median) | Methylation in Control | 4.27E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC6A4 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS1500 (cg18584905) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.47E+00 | Statistic Test | p-value: 8.69E-08; Z-score: 3.15E+00 | ||
|
Methylation in Case |
2.90E-01 (Median) | Methylation in Control | 1.98E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC6A4 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg25725890) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.65E+00 | Statistic Test | p-value: 3.04E-08; Z-score: 3.48E+00 | ||
|
Methylation in Case |
1.41E-01 (Median) | Methylation in Control | 8.58E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC6A4 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg27569822) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.85E+00 | Statistic Test | p-value: 5.39E-07; Z-score: 2.09E+00 | ||
|
Methylation in Case |
8.15E-02 (Median) | Methylation in Control | 4.40E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC6A4 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg10901968) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.12E+00 | Statistic Test | p-value: 1.15E-05; Z-score: 2.88E-01 | ||
|
Methylation in Case |
4.14E-02 (Median) | Methylation in Control | 3.71E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLC6A4 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg26741280) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.08E+00 | Statistic Test | p-value: 2.76E-05; Z-score: 3.92E-01 | ||
|
Methylation in Case |
1.63E-01 (Median) | Methylation in Control | 1.50E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 10 |
Methylation of SLC6A4 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
Body (cg18016139) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.91E+00 | Statistic Test | p-value: 3.59E-20; Z-score: -4.90E+00 | ||
|
Methylation in Case |
4.18E-01 (Median) | Methylation in Control | 7.96E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 11 |
Methylation of SLC6A4 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
Body (cg23516560) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.65E+00 | Statistic Test | p-value: 6.10E-17; Z-score: -5.34E+00 | ||
|
Methylation in Case |
4.48E-01 (Median) | Methylation in Control | 7.42E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 12 |
Methylation of SLC6A4 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
Body (cg16237262) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.44E+00 | Statistic Test | p-value: 1.13E-14; Z-score: -2.36E+00 | ||
|
Methylation in Case |
5.28E-01 (Median) | Methylation in Control | 7.62E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 13 |
Methylation of SLC6A4 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
Body (cg21136182) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.34E+00 | Statistic Test | p-value: 1.39E-13; Z-score: -6.81E+00 | ||
|
Methylation in Case |
6.38E-01 (Median) | Methylation in Control | 8.53E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 14 |
Methylation of SLC6A4 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
Body (cg20669572) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.18E+00 | Statistic Test | p-value: 2.36E-13; Z-score: 2.35E+00 | ||
|
Methylation in Case |
7.50E-01 (Median) | Methylation in Control | 6.38E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 15 |
Methylation of SLC6A4 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
Body (cg04661001) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.12E+00 | Statistic Test | p-value: 3.40E-13; Z-score: 2.00E+00 | ||
|
Methylation in Case |
8.01E-01 (Median) | Methylation in Control | 7.14E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 16 |
Methylation of SLC6A4 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
3'UTR (cg07479786) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.10E+00 | Statistic Test | p-value: 1.41E-09; Z-score: 2.26E+00 | ||
|
Methylation in Case |
8.26E-01 (Median) | Methylation in Control | 7.54E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 17 |
Methylation of SLC6A4 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
3'UTR (cg20592995) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.03E+00 | Statistic Test | p-value: 1.12E-04; Z-score: -7.46E-01 | ||
|
Methylation in Case |
7.93E-01 (Median) | Methylation in Control | 8.16E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
HIV infection |
9 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC6A4 in HIV infection | [ 8 ] | |||
|
Location |
5'UTR (cg05951817) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.07E+00 | Statistic Test | p-value: 2.17E-03; Z-score: -8.77E-01 | ||
|
Methylation in Case |
7.25E-01 (Median) | Methylation in Control | 7.77E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC6A4 in HIV infection | [ 8 ] | |||
|
Location |
5'UTR (cg01330016) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.01E+00 | Statistic Test | p-value: 2.51E-02; Z-score: 6.21E-01 | ||
|
Methylation in Case |
9.10E-01 (Median) | Methylation in Control | 8.97E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC6A4 in HIV infection | [ 8 ] | |||
|
Location |
TSS1500 (cg18584905) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.37E+00 | Statistic Test | p-value: 8.22E-06; Z-score: 2.00E+00 | ||
|
Methylation in Case |
1.99E-01 (Median) | Methylation in Control | 1.45E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC6A4 in HIV infection | [ 8 ] | |||
|
Location |
TSS200 (cg25725890) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.22E+00 | Statistic Test | p-value: 5.51E-05; Z-score: 1.19E+00 | ||
|
Methylation in Case |
9.44E-02 (Median) | Methylation in Control | 7.72E-02 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC6A4 in HIV infection | [ 8 ] | |||
|
Location |
TSS200 (cg27569822) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.47E+00 | Statistic Test | p-value: 4.33E-03; Z-score: 1.54E+00 | ||
|
Methylation in Case |
4.52E-02 (Median) | Methylation in Control | 3.08E-02 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC6A4 in HIV infection | [ 8 ] | |||
|
Location |
TSS200 (cg10901968) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.28E+00 | Statistic Test | p-value: 1.21E-02; Z-score: 1.17E+00 | ||
|
Methylation in Case |
4.32E-02 (Median) | Methylation in Control | 3.36E-02 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC6A4 in HIV infection | [ 8 ] | |||
|
Location |
TSS200 (cg26741280) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.04E+00 | Statistic Test | p-value: 2.82E-02; Z-score: 4.35E-01 | ||
|
Methylation in Case |
2.15E-01 (Median) | Methylation in Control | 2.07E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC6A4 in HIV infection | [ 8 ] | |||
|
Location |
Body (cg24984698) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.03E+00 | Statistic Test | p-value: 6.20E-04; Z-score: -1.03E+00 | ||
|
Methylation in Case |
8.27E-01 (Median) | Methylation in Control | 8.54E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLC6A4 in HIV infection | [ 8 ] | |||
|
Location |
3'UTR (cg20592995) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.04E+00 | Statistic Test | p-value: 2.96E-05; Z-score: -1.45E+00 | ||
|
Methylation in Case |
8.63E-01 (Median) | Methylation in Control | 8.96E-01 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Lung adenocarcinoma |
8 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC6A4 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
5'UTR (cg26126367) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.14E+00 | Statistic Test | p-value: 2.39E-03; Z-score: 1.57E+00 | ||
|
Methylation in Case |
7.91E-01 (Median) | Methylation in Control | 6.92E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC6A4 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
5'UTR (cg22584138) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.16E+00 | Statistic Test | p-value: 6.27E-03; Z-score: 1.96E+00 | ||
|
Methylation in Case |
5.52E-01 (Median) | Methylation in Control | 4.77E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC6A4 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
5'UTR (cg03363743) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.21E+00 | Statistic Test | p-value: 1.19E-02; Z-score: 1.64E+00 | ||
|
Methylation in Case |
3.78E-01 (Median) | Methylation in Control | 3.11E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC6A4 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
5'UTR (cg01330016) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.08E+00 | Statistic Test | p-value: 4.92E-02; Z-score: 1.03E+00 | ||
|
Methylation in Case |
8.04E-01 (Median) | Methylation in Control | 7.45E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC6A4 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
TSS1500 (cg18584905) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.31E+00 | Statistic Test | p-value: 1.65E-02; Z-score: 8.13E-01 | ||
|
Methylation in Case |
1.94E-01 (Median) | Methylation in Control | 1.47E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC6A4 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
TSS200 (cg27569822) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.34E+00 | Statistic Test | p-value: 1.80E-02; Z-score: 1.75E+00 | ||
|
Methylation in Case |
7.36E-02 (Median) | Methylation in Control | 5.50E-02 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLC6A4 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
TSS200 (cg10901968) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.18E+00 | Statistic Test | p-value: 1.91E-02; Z-score: 1.87E+00 | ||
|
Methylation in Case |
5.45E-02 (Median) | Methylation in Control | 4.62E-02 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLC6A4 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
3'UTR (cg20592995) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.03E+00 | Statistic Test | p-value: 1.69E-02; Z-score: -8.98E-01 | ||
|
Methylation in Case |
8.17E-01 (Median) | Methylation in Control | 8.40E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Papillary thyroid cancer |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC6A4 in papillary thyroid cancer | [ 10 ] | |||
|
Location |
5'UTR (cg26126367) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.00E+00 | Statistic Test | p-value: 2.18E-02; Z-score: -1.39E-01 | ||
|
Methylation in Case |
8.95E-01 (Median) | Methylation in Control | 8.97E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC6A4 in papillary thyroid cancer | [ 10 ] | |||
|
Location |
5'UTR (cg22584138) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.08E+00 | Statistic Test | p-value: 4.36E-02; Z-score: -4.74E-01 | ||
|
Methylation in Case |
3.02E-01 (Median) | Methylation in Control | 3.26E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC6A4 in papillary thyroid cancer | [ 10 ] | |||
|
Location |
Body (cg24984698) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.06E+00 | Statistic Test | p-value: 2.28E-10; Z-score: -1.49E+00 | ||
|
Methylation in Case |
7.79E-01 (Median) | Methylation in Control | 8.23E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Pancretic ductal adenocarcinoma |
6 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC6A4 in pancretic ductal adenocarcinoma | [ 11 ] | |||
|
Location |
TSS1500 (cg19313402) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.19E+00 | Statistic Test | p-value: 3.82E-03; Z-score: -9.26E-01 | ||
|
Methylation in Case |
5.91E-01 (Median) | Methylation in Control | 7.03E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC6A4 in pancretic ductal adenocarcinoma | [ 11 ] | |||
|
Location |
TSS200 (cg19788741) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 4.51E+00 | Statistic Test | p-value: 3.03E-15; Z-score: 2.93E+00 | ||
|
Methylation in Case |
2.84E-01 (Median) | Methylation in Control | 6.28E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC6A4 in pancretic ductal adenocarcinoma | [ 11 ] | |||
|
Location |
TSS200 (cg01385367) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.05E+00 | Statistic Test | p-value: 1.95E-02; Z-score: -3.53E-01 | ||
|
Methylation in Case |
4.90E-02 (Median) | Methylation in Control | 5.14E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLC6A4 in pancretic ductal adenocarcinoma | [ 11 ] | |||
|
Location |
1stExon (cg23357854) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.27E+00 | Statistic Test | p-value: 1.31E-05; Z-score: -1.35E+00 | ||
|
Methylation in Case |
1.35E-01 (Median) | Methylation in Control | 1.72E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLC6A4 in pancretic ductal adenocarcinoma | [ 11 ] | |||
|
Location |
Body (cg04764584) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.31E+00 | Statistic Test | p-value: 8.17E-12; Z-score: -2.16E+00 | ||
|
Methylation in Case |
3.36E-01 (Median) | Methylation in Control | 4.40E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLC6A4 in pancretic ductal adenocarcinoma | [ 11 ] | |||
|
Location |
Body (cg13603332) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.05E+00 | Statistic Test | p-value: 8.03E-05; Z-score: -2.68E-01 | ||
|
Methylation in Case |
5.00E-01 (Median) | Methylation in Control | 5.23E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Prostate cancer |
4 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC6A4 in prostate cancer | [ 12 ] | |||
|
Location |
TSS1500 (cg02511315) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.21E+00 | Statistic Test | p-value: 2.55E-04; Z-score: 6.08E+00 | ||
|
Methylation in Case |
8.70E-01 (Median) | Methylation in Control | 7.18E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC6A4 in prostate cancer | [ 12 ] | |||
|
Location |
TSS200 (cg25881850) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.66E+00 | Statistic Test | p-value: 3.52E-02; Z-score: 2.35E+00 | ||
|
Methylation in Case |
3.82E-01 (Median) | Methylation in Control | 2.30E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLC6A4 in prostate cancer | [ 12 ] | |||
|
Location |
Body (cg02595575) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 4.72E+00 | Statistic Test | p-value: 1.01E-02; Z-score: 5.03E+00 | ||
|
Methylation in Case |
4.16E-01 (Median) | Methylation in Control | 8.82E-02 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Moderate hypermethylation of SLC6A4 in prostate cancer than that in healthy individual | ||||
Studied Phenotype |
Prostate cancer [ICD-11:2C82] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 3.87E-10; Fold-change: 0.209304236; Z-score: 4.914445185 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
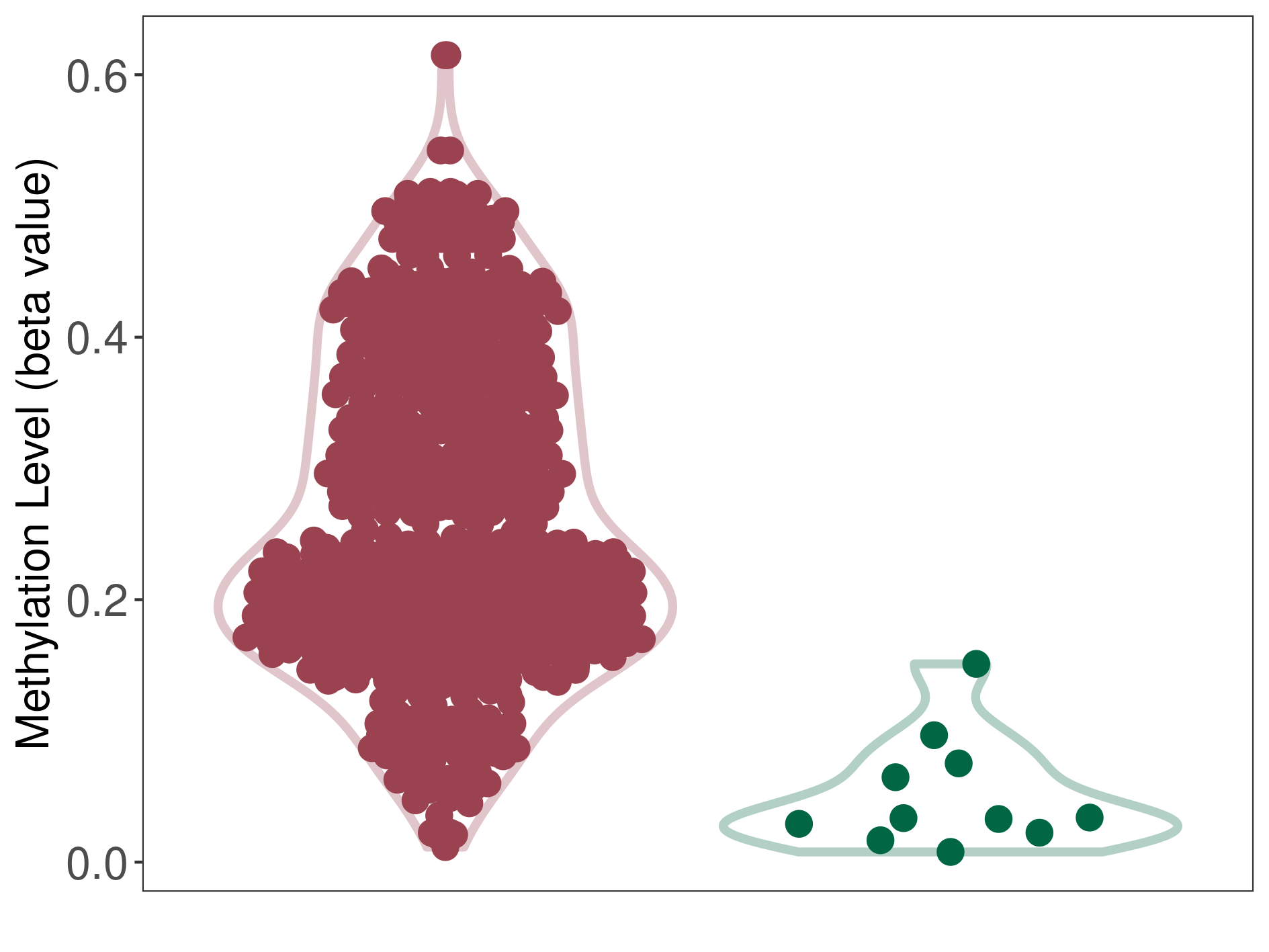
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Renal cell carcinoma |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC6A4 in clear cell renal cell carcinoma | [ 13 ] | |||
|
Location |
TSS200 (cg27569822) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.04E+00 | Statistic Test | p-value: 1.46E-02; Z-score: 2.90E-01 | ||
|
Methylation in Case |
2.48E-02 (Median) | Methylation in Control | 2.38E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLC6A4 in clear cell renal cell carcinoma | [ 13 ] | |||
|
Location |
TSS200 (cg26741280) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.04E+00 | Statistic Test | p-value: 3.64E-02; Z-score: 2.58E-01 | ||
|
Methylation in Case |
1.76E-01 (Median) | Methylation in Control | 1.69E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colon cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC6A4 in colon adenocarcinoma | [ 14 ] | |||
|
Location |
Body (cg02208504) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.13E+00 | Statistic Test | p-value: 2.17E-03; Z-score: -2.65E+00 | ||
|
Methylation in Case |
6.37E-01 (Median) | Methylation in Control | 7.21E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Systemic lupus erythematosus |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLC6A4 in systemic lupus erythematosus | [ 15 ] | |||
|
Location |
Body (cg24984698) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.01E+00 | Statistic Test | p-value: 4.31E-02; Z-score: -1.59E-01 | ||
|
Methylation in Case |
8.39E-01 (Median) | Methylation in Control | 8.47E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus [ ICD-11: 4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Depression |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Hypomethylation of SLC6A4 in depressive disorder | [ 19 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Bisulfite sequencing | ||
|
Studied Phenotype |
Depression [ ICD-11: 6A8Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Additional Notes |
DNA methylation level at the SLC6A4 promoter in the motherchild group, concordant with the depression diagnosis. | ||||
|
Acute myeloid leukemia |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Moderate hypermethylation of SLC6A4 in acute myeloid leukemia than that in healthy individual | ||||
Studied Phenotype |
Acute myeloid leukemia [ICD-11:2A60] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 1.89E-07; Fold-change: 0.210558189; Z-score: 1.582647806 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
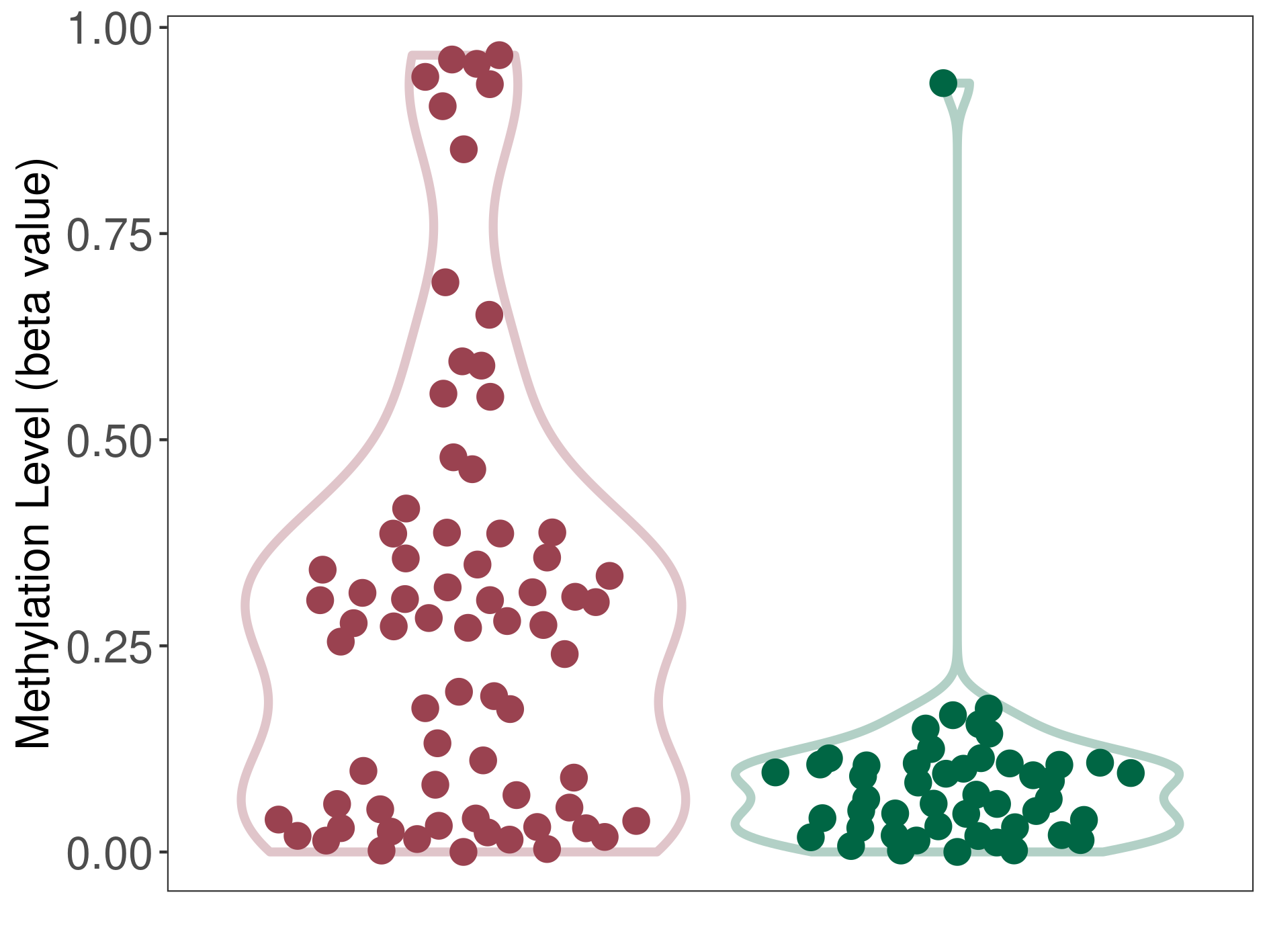
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Oligodendroglioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypermethylation of SLC6A4 in oligodendroglioma than that in healthy individual | ||||
Studied Phenotype |
Oligodendroglioma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 4.71E-35; Fold-change: 0.304047268; Z-score: 3.747691008 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
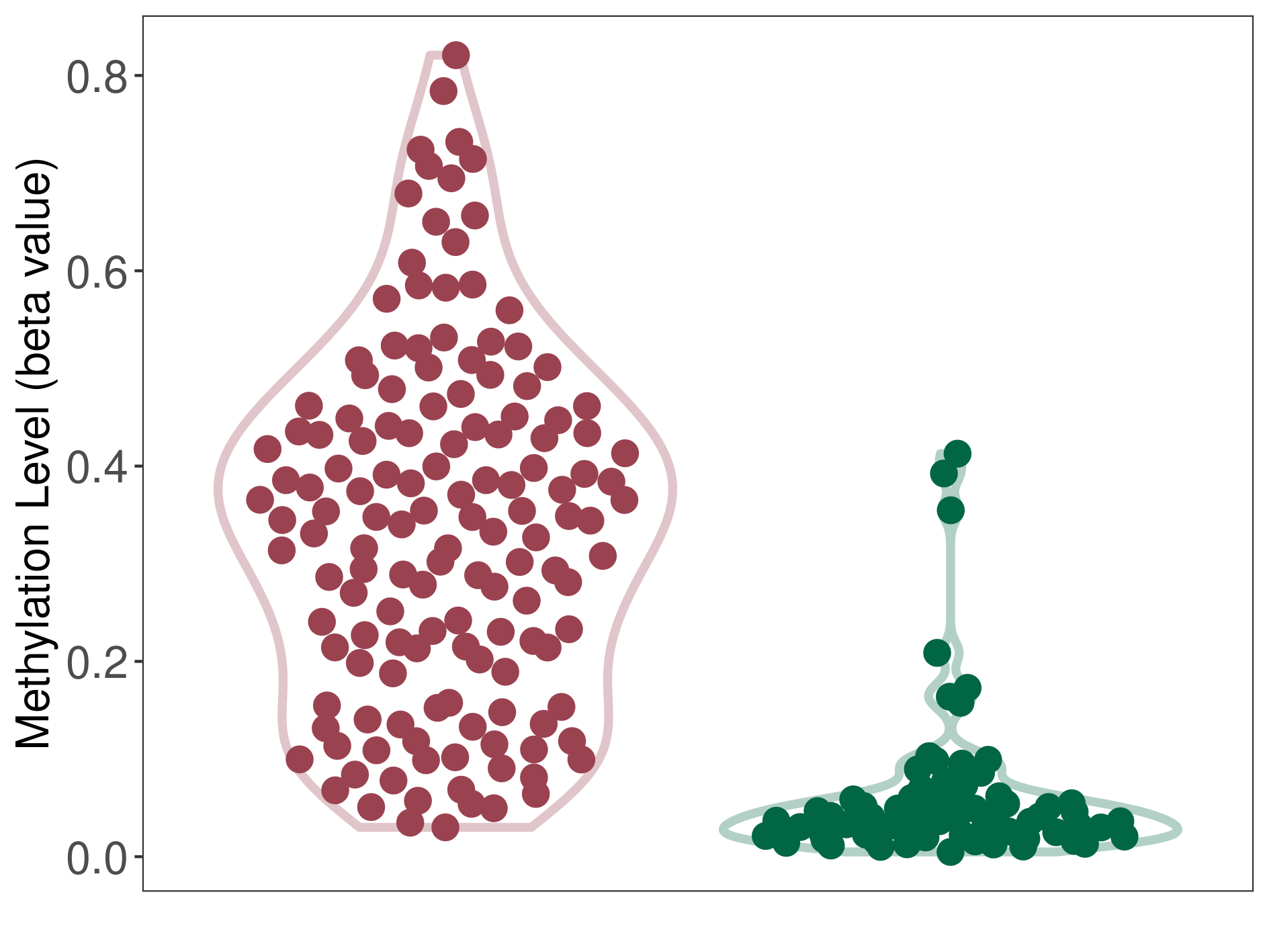
|
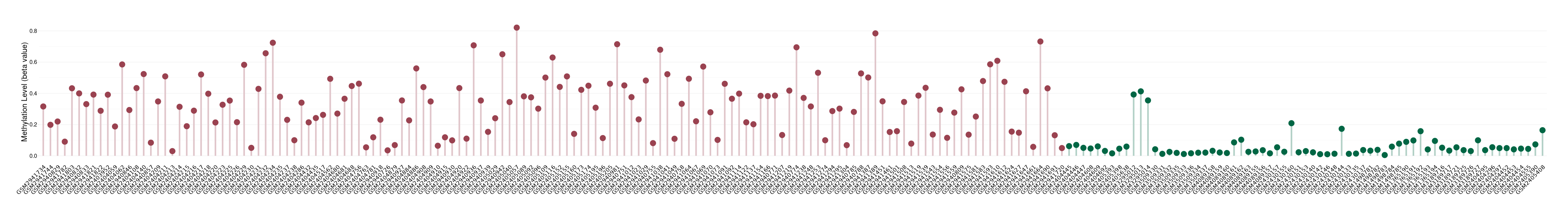 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Histone acetylation |
|||||
|
Colorectal cancer |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Hypoacetylation of SLC6A4 in epithelial colorectal adenocarcinoma (compare with butyrate-treatment counterpart cells) | [ 16 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes |
Down regulation of SLC6A4 | Experiment Method | Western Blot | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Human epithelial colorectal adenocarcinoma cells (Caco-2) | ||||
|
Additional Notes |
The SLC6A4 promoter 1 region appears to be specifically modulated by alterations in the acetylation (but not methylation) status of histone H3. | ||||
|
Epigenetic Phenomenon 2 |
Hypoacetylation of SLC6A4 in epithelial colorectal adenocarcinoma (compare with butyrate-treatment counterpart cells) | [ 16 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes |
Down regulation of SLC6A4 | Experiment Method | Western Blot | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Human epithelial colorectal adenocarcinoma cells (Caco-2) | ||||
|
Additional Notes |
The SLC6A4 promoter 1 region appears to be specifically modulated by alterations in the acetylation (but not methylation) status of histone H4. | ||||
|
Several types of cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Hypoacetylation of SLC6A4 in several types of cancer (compare with trichostatin A treatment counterpart cells) | [ 17 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes |
Down regulation of SLC6A4 | Experiment Method | Western Blot | ||
|
Studied Phenotype |
Several types of cancer [ ICD-11: 2A00-2F9Z] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
SLC6A4 gene is epigenetically downregulated by HDAC1 in several types of cancer. | ||||
|
Neuroblastoma cells |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Hypoacetylation of SLC6A4 in neuroblastoma cells (compare with TSA-treatment counterpart cells) | [ 18 ] | |||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes |
Down regulation of SLC6A4 | Experiment Method | Western Blot | ||
|
Studied Phenotype |
Neuroblastoma cells | ||||
|
Experimental Material |
Human neuroblastoma cell line (SK-NF-I) | ||||
|
Additional Notes |
Dramatic upregulation of dopamine and serotonin transporter expression by inhibition of histone deacetylases. | ||||
|
microRNA |
|||||
|
Unclear Phenotype |
68 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
miR-106a directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-106a | miRNA Mature ID | miR-106a-5p | ||
|
miRNA Sequence |
AAAAGUGCUUACAGUGCAGGUAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 2 |
miR-106b directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-106b | miRNA Mature ID | miR-106b-5p | ||
|
miRNA Sequence |
UAAAGUGCUGACAGUGCAGAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 3 |
miR-1184 directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-1184 | miRNA Mature ID | miR-1184 | ||
|
miRNA Sequence |
CCUGCAGCGACUUGAUGGCUUCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 4 |
miR-1205 directly targets SLC6A4 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-1205 | miRNA Mature ID | miR-1205 | ||
|
miRNA Sequence |
UCUGCAGGGUUUGCUUUGAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 5 |
miR-1237 directly targets SLC6A4 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-1237 | miRNA Mature ID | miR-1237-3p | ||
|
miRNA Sequence |
UCCUUCUGCUCCGUCCCCCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 6 |
miR-1264 directly targets SLC6A4 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-1264 | miRNA Mature ID | miR-1264 | ||
|
miRNA Sequence |
CAAGUCUUAUUUGAGCACCUGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 7 |
miR-1343 directly targets SLC6A4 | [ 22 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-1343 | miRNA Mature ID | miR-1343-5p | ||
|
miRNA Sequence |
UGGGGAGCGGCCCCCGGGUGGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 8 |
miR-141 directly targets SLC6A4 | [ 22 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-141 | miRNA Mature ID | miR-141-5p | ||
|
miRNA Sequence |
CAUCUUCCAGUACAGUGUUGGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 9 |
miR-142 directly targets SLC6A4 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-142 | miRNA Mature ID | miR-142-5p | ||
|
miRNA Sequence |
CAUAAAGUAGAAAGCACUACU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 10 |
miR-150 directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-150 | miRNA Mature ID | miR-150-5p | ||
|
miRNA Sequence |
UCUCCCAACCCUUGUACCAGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 11 |
miR-17 directly targets SLC6A4 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-17 | miRNA Mature ID | miR-17-3p | ||
|
miRNA Sequence |
ACUGCAGUGAAGGCACUUGUAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 12 |
miR-17 directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-17 | miRNA Mature ID | miR-17-5p | ||
|
miRNA Sequence |
CAAAGUGCUUACAGUGCAGGUAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 13 |
miR-186 directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-186 | miRNA Mature ID | miR-186-3p | ||
|
miRNA Sequence |
GCCCAAAGGUGAAUUUUUUGGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 14 |
miR-20a directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-20a | miRNA Mature ID | miR-20a-5p | ||
|
miRNA Sequence |
UAAAGUGCUUAUAGUGCAGGUAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 15 |
miR-20b directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-20b | miRNA Mature ID | miR-20b-5p | ||
|
miRNA Sequence |
CAAAGUGCUCAUAGUGCAGGUAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 16 |
miR-2467 directly targets SLC6A4 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-2467 | miRNA Mature ID | miR-2467-3p | ||
|
miRNA Sequence |
AGCAGAGGCAGAGAGGCUCAGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 17 |
miR-302a directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-302a | miRNA Mature ID | miR-302a-3p | ||
|
miRNA Sequence |
UAAGUGCUUCCAUGUUUUGGUGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 18 |
miR-302b directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-302b | miRNA Mature ID | miR-302b-3p | ||
|
miRNA Sequence |
UAAGUGCUUCCAUGUUUUAGUAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 19 |
miR-302c directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-302c | miRNA Mature ID | miR-302c-3p | ||
|
miRNA Sequence |
UAAGUGCUUCCAUGUUUCAGUGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 20 |
miR-302d directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-302d | miRNA Mature ID | miR-302d-3p | ||
|
miRNA Sequence |
UAAGUGCUUCCAUGUUUGAGUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 21 |
miR-302e directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-302e | miRNA Mature ID | miR-302e | ||
|
miRNA Sequence |
UAAGUGCUUCCAUGCUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 22 |
miR-3130 directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-3130 | miRNA Mature ID | miR-3130-3p | ||
|
miRNA Sequence |
GCUGCACCGGAGACUGGGUAA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 23 |
miR-3158 directly targets SLC6A4 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-3158 | miRNA Mature ID | miR-3158-5p | ||
|
miRNA Sequence |
CCUGCAGAGAGGAAGCCCUUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 24 |
miR-3175 directly targets SLC6A4 | [ 22 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3175 | miRNA Mature ID | miR-3175 | ||
|
miRNA Sequence |
CGGGGAGAGAACGCAGUGACGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 25 |
miR-335 directly targets SLC6A4 | [ 23 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Microarray | ||
|
miRNA Stemloop ID |
miR-335 | miRNA Mature ID | miR-335-5p | ||
|
miRNA Sequence |
UCAAGAGCAAUAACGAAAAAUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 26 |
miR-3678 directly targets SLC6A4 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-3678 | miRNA Mature ID | miR-3678-3p | ||
|
miRNA Sequence |
CUGCAGAGUUUGUACGGACCGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 27 |
miR-372 directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-372 | miRNA Mature ID | miR-372-3p | ||
|
miRNA Sequence |
AAAGUGCUGCGACAUUUGAGCGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 28 |
miR-373 directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-373 | miRNA Mature ID | miR-373-3p | ||
|
miRNA Sequence |
GAAGUGCUUCGAUUUUGGGGUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 29 |
miR-3934 directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-3934 | miRNA Mature ID | miR-3934-5p | ||
|
miRNA Sequence |
UCAGGUGUGGAAACUGAGGCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 30 |
miR-4287 directly targets SLC6A4 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4287 | miRNA Mature ID | miR-4287 | ||
|
miRNA Sequence |
UCUCCCUUGAGGGCACUUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 31 |
miR-4418 directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4418 | miRNA Mature ID | miR-4418 | ||
|
miRNA Sequence |
CACUGCAGGACUCAGCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 32 |
miR-4685 directly targets SLC6A4 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4685 | miRNA Mature ID | miR-4685-3p | ||
|
miRNA Sequence |
UCUCCCUUCCUGCCCUGGCUAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 33 |
miR-4691 directly targets SLC6A4 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4691 | miRNA Mature ID | miR-4691-5p | ||
|
miRNA Sequence |
GUCCUCCAGGCCAUGAGCUGCGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 34 |
miR-4731 directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4731 | miRNA Mature ID | miR-4731-5p | ||
|
miRNA Sequence |
UGCUGGGGGCCACAUGAGUGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 35 |
miR-491 directly targets SLC6A4 | [ 22 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-491 | miRNA Mature ID | miR-491-5p | ||
|
miRNA Sequence |
AGUGGGGAACCCUUCCAUGAGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 36 |
miR-5088 directly targets SLC6A4 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-5088 | miRNA Mature ID | miR-5088-3p | ||
|
miRNA Sequence |
UCCCUUCUUCCUGGGCCCUCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 37 |
miR-5089 directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-5089 | miRNA Mature ID | miR-5089-5p | ||
|
miRNA Sequence |
GUGGGAUUUCUGAGUAGCAUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 38 |
miR-509 directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-509 | miRNA Mature ID | miR-509-5p | ||
|
miRNA Sequence |
UACUGCAGACAGUGGCAAUCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 39 |
miR-512 directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-512 | miRNA Mature ID | miR-512-3p | ||
|
miRNA Sequence |
AAGUGCUGUCAUAGCUGAGGUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 40 |
miR-519d directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-519d | miRNA Mature ID | miR-519d-3p | ||
|
miRNA Sequence |
CAAAGUGCCUCCCUUUAGAGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 41 |
miR-520a directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-520a | miRNA Mature ID | miR-520a-3p | ||
|
miRNA Sequence |
AAAGUGCUUCCCUUUGGACUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 42 |
miR-520c directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-520c | miRNA Mature ID | miR-520c-3p | ||
|
miRNA Sequence |
AAAGUGCUUCCUUUUAGAGGGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 43 |
miR-520d directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-520d | miRNA Mature ID | miR-520d-3p | ||
|
miRNA Sequence |
AAAGUGCUUCUCUUUGGUGGGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 44 |
miR-520g directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-520g | miRNA Mature ID | miR-520g-3p | ||
|
miRNA Sequence |
ACAAAGUGCUUCCCUUUAGAGUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 45 |
miR-520h directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-520h | miRNA Mature ID | miR-520h | ||
|
miRNA Sequence |
ACAAAGUGCUUCCCUUUAGAGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 46 |
miR-526b directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-526b | miRNA Mature ID | miR-526b-3p | ||
|
miRNA Sequence |
GAAAGUGCUUCCUUUUAGAGGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 47 |
miR-544a directly targets SLC6A4 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-544a | miRNA Mature ID | miR-544a | ||
|
miRNA Sequence |
AUUCUGCAUUUUUAGCAAGUUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 48 |
miR-552 directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-552 | miRNA Mature ID | miR-552-3p | ||
|
miRNA Sequence |
AACAGGUGACUGGUUAGACAA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 49 |
miR-5589 directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-5589 | miRNA Mature ID | miR-5589-5p | ||
|
miRNA Sequence |
GGCUGGGUGCUCUUGUGCAGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 50 |
miR-5590 directly targets SLC6A4 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-5590 | miRNA Mature ID | miR-5590-3p | ||
|
miRNA Sequence |
AAUAAAGUUCAUGUAUGGCAA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 51 |
miR-5702 directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-5702 | miRNA Mature ID | miR-5702 | ||
|
miRNA Sequence |
UGAGUCAGCAACAUAUCCCAUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 52 |
miR-619 directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-619 | miRNA Mature ID | miR-619-5p | ||
|
miRNA Sequence |
GCUGGGAUUACAGGCAUGAGCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 53 |
miR-6504 directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6504 | miRNA Mature ID | miR-6504-3p | ||
|
miRNA Sequence |
CAUUACAGCACAGCCAUUCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 54 |
miR-6506 directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6506 | miRNA Mature ID | miR-6506-5p | ||
|
miRNA Sequence |
ACUGGGAUGUCACUGAAUAUGGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 55 |
miR-6507 directly targets SLC6A4 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6507 | miRNA Mature ID | miR-6507-3p | ||
|
miRNA Sequence |
CAAAGUCCUUCCUAUUUUUCCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 56 |
miR-6749 directly targets SLC6A4 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6749 | miRNA Mature ID | miR-6749-3p | ||
|
miRNA Sequence |
CUCCUCCCCUGCCUGGCCCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 57 |
miR-6756 directly targets SLC6A4 | [ 22 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6756 | miRNA Mature ID | miR-6756-5p | ||
|
miRNA Sequence |
AGGGUGGGGCUGGAGGUGGGGCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 58 |
miR-6763 directly targets SLC6A4 | [ 22 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6763 | miRNA Mature ID | miR-6763-5p | ||
|
miRNA Sequence |
CUGGGGAGUGGCUGGGGAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 59 |
miR-6766 directly targets SLC6A4 | [ 22 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6766 | miRNA Mature ID | miR-6766-5p | ||
|
miRNA Sequence |
CGGGUGGGAGCAGAUCUUAUUGAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 60 |
miR-6792 directly targets SLC6A4 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6792 | miRNA Mature ID | miR-6792-3p | ||
|
miRNA Sequence |
CUCCUCCACAGCCCCUGCUCAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 61 |
miR-6825 directly targets SLC6A4 | [ 22 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6825 | miRNA Mature ID | miR-6825-5p | ||
|
miRNA Sequence |
UGGGGAGGUGUGGAGUCAGCAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 62 |
miR-6878 directly targets SLC6A4 | [ 22 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6878 | miRNA Mature ID | miR-6878-5p | ||
|
miRNA Sequence |
AGGGAGAAAGCUAGAAGCUGAAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 63 |
miR-6890 directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6890 | miRNA Mature ID | miR-6890-3p | ||
|
miRNA Sequence |
CCACUGCCUAUGCCCCACAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 64 |
miR-7151 directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-7151 | miRNA Mature ID | miR-7151-3p | ||
|
miRNA Sequence |
CUACAGGCUGGAAUGGGCUCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 65 |
miR-7156 directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-7156 | miRNA Mature ID | miR-7156-3p | ||
|
miRNA Sequence |
CUGCAGCCACUUGGGGAACUGGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 66 |
miR-93 directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-93 | miRNA Mature ID | miR-93-5p | ||
|
miRNA Sequence |
CAAAGUGCUGUUCGUGCAGGUAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 67 |
miR-937 directly targets SLC6A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-937 | miRNA Mature ID | miR-937-5p | ||
|
miRNA Sequence |
GUGAGUCAGGGUGGGGCUGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 68 |
miR-939 directly targets SLC6A4 | [ 22 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-939 | miRNA Mature ID | miR-939-5p | ||
|
miRNA Sequence |
UGGGGAGCUGAGGCUCUGGGGGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.
