Detail Information of Epigenetic Regulations
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0503 Transporter Info | ||||
| Gene Name | SLCO4C1 | ||||
| Transporter Name | Organic anion transporting polypeptide 4C1 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| Epigenetic Regulations of This DT (EGR) | |||||
|---|---|---|---|---|---|
|
Methylation |
|||||
|
Pancretic ductal adenocarcinoma |
9 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLCO4C1 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
5'UTR (cg03721454) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.04E+00 | Statistic Test | p-value: 6.36E-03; Z-score: 4.25E-01 | ||
|
Methylation in Case |
7.18E-01 (Median) | Methylation in Control | 6.91E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLCO4C1 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg14482308) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.54E+00 | Statistic Test | p-value: 2.66E-14; Z-score: 2.24E+00 | ||
|
Methylation in Case |
4.32E-01 (Median) | Methylation in Control | 2.80E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLCO4C1 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg26360695) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.19E+00 | Statistic Test | p-value: 3.39E-03; Z-score: -8.12E-01 | ||
|
Methylation in Case |
1.16E-01 (Median) | Methylation in Control | 1.39E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLCO4C1 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg23375719) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.19E+00 | Statistic Test | p-value: 6.26E-03; Z-score: -4.47E-01 | ||
|
Methylation in Case |
1.17E-01 (Median) | Methylation in Control | 1.39E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLCO4C1 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg17221813) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.08E+00 | Statistic Test | p-value: 3.56E-04; Z-score: 8.90E-01 | ||
|
Methylation in Case |
7.88E-01 (Median) | Methylation in Control | 7.29E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLCO4C1 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg10117207) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.18E+00 | Statistic Test | p-value: 1.17E-03; Z-score: 1.09E+00 | ||
|
Methylation in Case |
8.56E-01 (Median) | Methylation in Control | 7.28E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLCO4C1 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg04070804) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.04E+00 | Statistic Test | p-value: 4.54E-03; Z-score: 9.05E-01 | ||
|
Methylation in Case |
8.74E-01 (Median) | Methylation in Control | 8.44E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLCO4C1 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg18312989) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.07E+00 | Statistic Test | p-value: 1.27E-02; Z-score: -6.01E-01 | ||
|
Methylation in Case |
5.07E-01 (Median) | Methylation in Control | 5.45E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLCO4C1 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
3'UTR (cg14022794) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.10E+00 | Statistic Test | p-value: 9.93E-04; Z-score: -9.85E-01 | ||
|
Methylation in Case |
3.45E-01 (Median) | Methylation in Control | 3.80E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Bladder cancer |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLCO4C1 in bladder cancer | [ 2 ] | |||
|
Location |
TSS1500 (cg12864568) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -2.36E+00 | Statistic Test | p-value: 2.76E-06; Z-score: -5.46E+00 | ||
|
Methylation in Case |
1.13E-01 (Median) | Methylation in Control | 2.67E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLCO4C1 in bladder cancer | [ 2 ] | |||
|
Location |
TSS1500 (cg24036612) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.05E+00 | Statistic Test | p-value: 2.72E-03; Z-score: -4.69E+00 | ||
|
Methylation in Case |
8.20E-01 (Median) | Methylation in Control | 8.65E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLCO4C1 in bladder cancer | [ 2 ] | |||
|
Location |
TSS1500 (cg19774478) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.86E+00 | Statistic Test | p-value: 1.24E-02; Z-score: -2.34E+00 | ||
|
Methylation in Case |
7.08E-02 (Median) | Methylation in Control | 1.32E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Breast cancer |
9 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLCO4C1 in breast cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg19774478) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.01E+00 | Statistic Test | p-value: 2.83E-03; Z-score: -2.13E-02 | ||
|
Methylation in Case |
7.56E-02 (Median) | Methylation in Control | 7.62E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLCO4C1 in breast cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg24036612) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.03E+00 | Statistic Test | p-value: 3.08E-02; Z-score: -7.04E-01 | ||
|
Methylation in Case |
8.44E-01 (Median) | Methylation in Control | 8.69E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLCO4C1 in breast cancer | [ 3 ] | |||
|
Location |
TSS200 (cg19788741) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.09E+00 | Statistic Test | p-value: 5.54E-03; Z-score: -1.40E-01 | ||
|
Methylation in Case |
3.24E-02 (Median) | Methylation in Control | 3.53E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLCO4C1 in breast cancer | [ 3 ] | |||
|
Location |
TSS200 (cg11267955) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.12E+00 | Statistic Test | p-value: 6.44E-03; Z-score: -4.26E-01 | ||
|
Methylation in Case |
5.47E-02 (Median) | Methylation in Control | 6.15E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLCO4C1 in breast cancer | [ 3 ] | |||
|
Location |
TSS200 (cg04633600) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.11E+00 | Statistic Test | p-value: 1.58E-02; Z-score: 3.03E-01 | ||
|
Methylation in Case |
4.91E-02 (Median) | Methylation in Control | 4.41E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLCO4C1 in breast cancer | [ 3 ] | |||
|
Location |
TSS200 (cg04621020) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.09E+00 | Statistic Test | p-value: 2.01E-02; Z-score: -4.02E-01 | ||
|
Methylation in Case |
5.96E-02 (Median) | Methylation in Control | 6.52E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLCO4C1 in breast cancer | [ 3 ] | |||
|
Location |
TSS200 (cg06480736) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.06E+00 | Statistic Test | p-value: 2.01E-02; Z-score: -2.38E-01 | ||
|
Methylation in Case |
6.40E-02 (Median) | Methylation in Control | 6.79E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLCO4C1 in breast cancer | [ 3 ] | |||
|
Location |
Body (cg23318786) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.10E+00 | Statistic Test | p-value: 1.39E-02; Z-score: 2.18E-01 | ||
|
Methylation in Case |
2.73E-02 (Median) | Methylation in Control | 2.49E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLCO4C1 in breast cancer | [ 3 ] | |||
|
Location |
3'UTR (cg00630480) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.09E+00 | Statistic Test | p-value: 7.63E-07; Z-score: 9.00E-01 | ||
|
Methylation in Case |
6.03E-01 (Median) | Methylation in Control | 5.52E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Renal cell carcinoma |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLCO4C1 in clear cell renal cell carcinoma | [ 4 ] | |||
|
Location |
TSS1500 (cg12864568) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.66E+00 | Statistic Test | p-value: 3.80E-02; Z-score: -1.81E+00 | ||
|
Methylation in Case |
1.15E-01 (Median) | Methylation in Control | 1.91E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLCO4C1 in clear cell renal cell carcinoma | [ 4 ] | |||
|
Location |
TSS200 (cg22149516) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.46E+00 | Statistic Test | p-value: 1.17E-02; Z-score: -1.17E+00 | ||
|
Methylation in Case |
6.30E-02 (Median) | Methylation in Control | 9.18E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLCO4C1 in clear cell renal cell carcinoma | [ 4 ] | |||
|
Location |
Body (cg23318786) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.04E+00 | Statistic Test | p-value: 1.17E-03; Z-score: 3.23E-01 | ||
|
Methylation in Case |
1.82E-02 (Median) | Methylation in Control | 1.74E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colon cancer |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLCO4C1 in colon adenocarcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg03586474) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.77E+00 | Statistic Test | p-value: 1.80E-03; Z-score: 3.56E+00 | ||
|
Methylation in Case |
2.33E-01 (Median) | Methylation in Control | 1.32E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLCO4C1 in colon adenocarcinoma | [ 5 ] | |||
|
Location |
Body (cg03366891) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.10E+00 | Statistic Test | p-value: 2.16E-04; Z-score: -3.03E+00 | ||
|
Methylation in Case |
7.22E-01 (Median) | Methylation in Control | 7.96E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colorectal cancer |
13 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLCO4C1 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS1500 (cg19774478) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.33E+00 | Statistic Test | p-value: 3.68E-05; Z-score: 1.02E+00 | ||
|
Methylation in Case |
2.53E-01 (Median) | Methylation in Control | 1.91E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLCO4C1 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS1500 (cg12864568) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.56E+00 | Statistic Test | p-value: 1.26E-04; Z-score: 1.62E+00 | ||
|
Methylation in Case |
4.54E-01 (Median) | Methylation in Control | 2.91E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLCO4C1 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS200 (cg04621020) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 5.44E+00 | Statistic Test | p-value: 4.80E-13; Z-score: 6.66E+00 | ||
|
Methylation in Case |
5.50E-01 (Median) | Methylation in Control | 1.01E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLCO4C1 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS200 (cg06480736) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 6.46E+00 | Statistic Test | p-value: 2.11E-12; Z-score: 7.33E+00 | ||
|
Methylation in Case |
3.71E-01 (Median) | Methylation in Control | 5.74E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLCO4C1 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS200 (cg11267955) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 8.40E+00 | Statistic Test | p-value: 3.88E-12; Z-score: 7.65E+00 | ||
|
Methylation in Case |
4.00E-01 (Median) | Methylation in Control | 4.76E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLCO4C1 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS200 (cg19788741) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.18E+01 | Statistic Test | p-value: 4.49E-12; Z-score: 6.05E+00 | ||
|
Methylation in Case |
4.42E-01 (Median) | Methylation in Control | 3.73E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLCO4C1 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS200 (cg22149516) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 3.42E+00 | Statistic Test | p-value: 4.74E-12; Z-score: 4.35E+00 | ||
|
Methylation in Case |
5.86E-01 (Median) | Methylation in Control | 1.71E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLCO4C1 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS200 (cg11009817) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 3.59E+00 | Statistic Test | p-value: 1.05E-11; Z-score: 5.09E+00 | ||
|
Methylation in Case |
4.04E-01 (Median) | Methylation in Control | 1.12E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLCO4C1 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS200 (cg04633600) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 4.17E+00 | Statistic Test | p-value: 2.90E-10; Z-score: 6.04E+00 | ||
|
Methylation in Case |
2.08E-01 (Median) | Methylation in Control | 5.00E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 10 |
Methylation of SLCO4C1 in colorectal cancer | [ 6 ] | |||
|
Location |
1stExon (cg00056676) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.44E+00 | Statistic Test | p-value: 2.86E-08; Z-score: 2.44E+00 | ||
|
Methylation in Case |
1.69E-01 (Median) | Methylation in Control | 1.17E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 11 |
Methylation of SLCO4C1 in colorectal cancer | [ 6 ] | |||
|
Location |
Body (cg23318786) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 2.37E+00 | Statistic Test | p-value: 1.46E-08; Z-score: 5.96E+00 | ||
|
Methylation in Case |
4.58E-02 (Median) | Methylation in Control | 1.93E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 12 |
Methylation of SLCO4C1 in colorectal cancer | [ 6 ] | |||
|
Location |
3'UTR (cg00630480) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.06E+00 | Statistic Test | p-value: 8.49E-05; Z-score: -1.23E+00 | ||
|
Methylation in Case |
7.04E-01 (Median) | Methylation in Control | 7.47E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Hepatocellular carcinoma |
9 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLCO4C1 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS1500 (cg19774478) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.33E+00 | Statistic Test | p-value: 6.88E-06; Z-score: 1.35E+00 | ||
|
Methylation in Case |
1.32E-01 (Median) | Methylation in Control | 9.96E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLCO4C1 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS1500 (cg24036612) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.04E+00 | Statistic Test | p-value: 4.53E-05; Z-score: -6.01E-01 | ||
|
Methylation in Case |
7.70E-01 (Median) | Methylation in Control | 8.03E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLCO4C1 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg11267955) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 3.01E+00 | Statistic Test | p-value: 3.07E-09; Z-score: 4.29E+00 | ||
|
Methylation in Case |
1.58E-01 (Median) | Methylation in Control | 5.25E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLCO4C1 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg04621020) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 2.43E+00 | Statistic Test | p-value: 5.14E-09; Z-score: 5.54E+00 | ||
|
Methylation in Case |
1.60E-01 (Median) | Methylation in Control | 6.57E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLCO4C1 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg06480736) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.91E+00 | Statistic Test | p-value: 6.97E-09; Z-score: 2.44E+00 | ||
|
Methylation in Case |
1.33E-01 (Median) | Methylation in Control | 6.97E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLCO4C1 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg11009817) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.64E+00 | Statistic Test | p-value: 2.20E-07; Z-score: 3.38E+00 | ||
|
Methylation in Case |
1.03E-01 (Median) | Methylation in Control | 6.31E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLCO4C1 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg04633600) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.26E+00 | Statistic Test | p-value: 2.35E-06; Z-score: 6.92E-01 | ||
|
Methylation in Case |
6.60E-02 (Median) | Methylation in Control | 5.24E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of SLCO4C1 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
1stExon (cg00056676) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.03E+00 | Statistic Test | p-value: 2.74E-02; Z-score: 1.70E-01 | ||
|
Methylation in Case |
8.40E-02 (Median) | Methylation in Control | 8.17E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of SLCO4C1 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
Body (cg09647867) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.36E+00 | Statistic Test | p-value: 3.68E-15; Z-score: -4.63E+00 | ||
|
Methylation in Case |
4.92E-01 (Median) | Methylation in Control | 6.69E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
HIV infection |
7 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLCO4C1 in HIV infection | [ 8 ] | |||
|
Location |
TSS1500 (cg19774478) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.18E+00 | Statistic Test | p-value: 4.15E-03; Z-score: 6.51E-01 | ||
|
Methylation in Case |
5.83E-02 (Median) | Methylation in Control | 4.95E-02 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLCO4C1 in HIV infection | [ 8 ] | |||
|
Location |
TSS1500 (cg12864568) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.32E+00 | Statistic Test | p-value: 1.88E-02; Z-score: 1.07E+00 | ||
|
Methylation in Case |
8.71E-02 (Median) | Methylation in Control | 6.59E-02 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLCO4C1 in HIV infection | [ 8 ] | |||
|
Location |
TSS200 (cg19788741) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.32E+00 | Statistic Test | p-value: 4.15E-03; Z-score: 1.06E+00 | ||
|
Methylation in Case |
2.92E-02 (Median) | Methylation in Control | 2.20E-02 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of SLCO4C1 in HIV infection | [ 8 ] | |||
|
Location |
TSS200 (cg11009817) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.07E+00 | Statistic Test | p-value: 2.66E-02; Z-score: 3.03E-01 | ||
|
Methylation in Case |
4.28E-02 (Median) | Methylation in Control | 4.00E-02 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of SLCO4C1 in HIV infection | [ 8 ] | |||
|
Location |
TSS200 (cg06480736) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.12E+00 | Statistic Test | p-value: 4.39E-02; Z-score: 4.27E-01 | ||
|
Methylation in Case |
5.19E-02 (Median) | Methylation in Control | 4.64E-02 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of SLCO4C1 in HIV infection | [ 8 ] | |||
|
Location |
TSS200 (cg04633600) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.12E+00 | Statistic Test | p-value: 4.62E-02; Z-score: 5.51E-01 | ||
|
Methylation in Case |
8.50E-02 (Median) | Methylation in Control | 7.62E-02 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of SLCO4C1 in HIV infection | [ 8 ] | |||
|
Location |
Body (cg23318786) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.40E+00 | Statistic Test | p-value: 2.49E-03; Z-score: 6.90E-01 | ||
|
Methylation in Case |
1.99E-02 (Median) | Methylation in Control | 1.42E-02 (Median) | ||
|
Studied Phenotype |
HIV infection [ ICD-11: 1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Papillary thyroid cancer |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLCO4C1 in papillary thyroid cancer | [ 9 ] | |||
|
Location |
TSS1500 (cg24036612) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.02E+00 | Statistic Test | p-value: 2.27E-04; Z-score: -7.93E-01 | ||
|
Methylation in Case |
8.59E-01 (Median) | Methylation in Control | 8.76E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLCO4C1 in papillary thyroid cancer | [ 9 ] | |||
|
Location |
TSS200 (cg11009817) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.26E+00 | Statistic Test | p-value: 4.73E-02; Z-score: -1.03E+00 | ||
|
Methylation in Case |
6.82E-02 (Median) | Methylation in Control | 8.58E-02 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLCO4C1 in papillary thyroid cancer | [ 9 ] | |||
|
Location |
Body (cg23318786) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.04E+00 | Statistic Test | p-value: 8.47E-03; Z-score: 2.63E-01 | ||
|
Methylation in Case |
5.49E-02 (Median) | Methylation in Control | 5.30E-02 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Prostate cancer |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLCO4C1 in prostate cancer | [ 10 ] | |||
|
Location |
TSS1500 (cg27365825) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 3.05E+00 | Statistic Test | p-value: 3.12E-04; Z-score: 1.13E+01 | ||
|
Methylation in Case |
6.29E-01 (Median) | Methylation in Control | 2.06E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
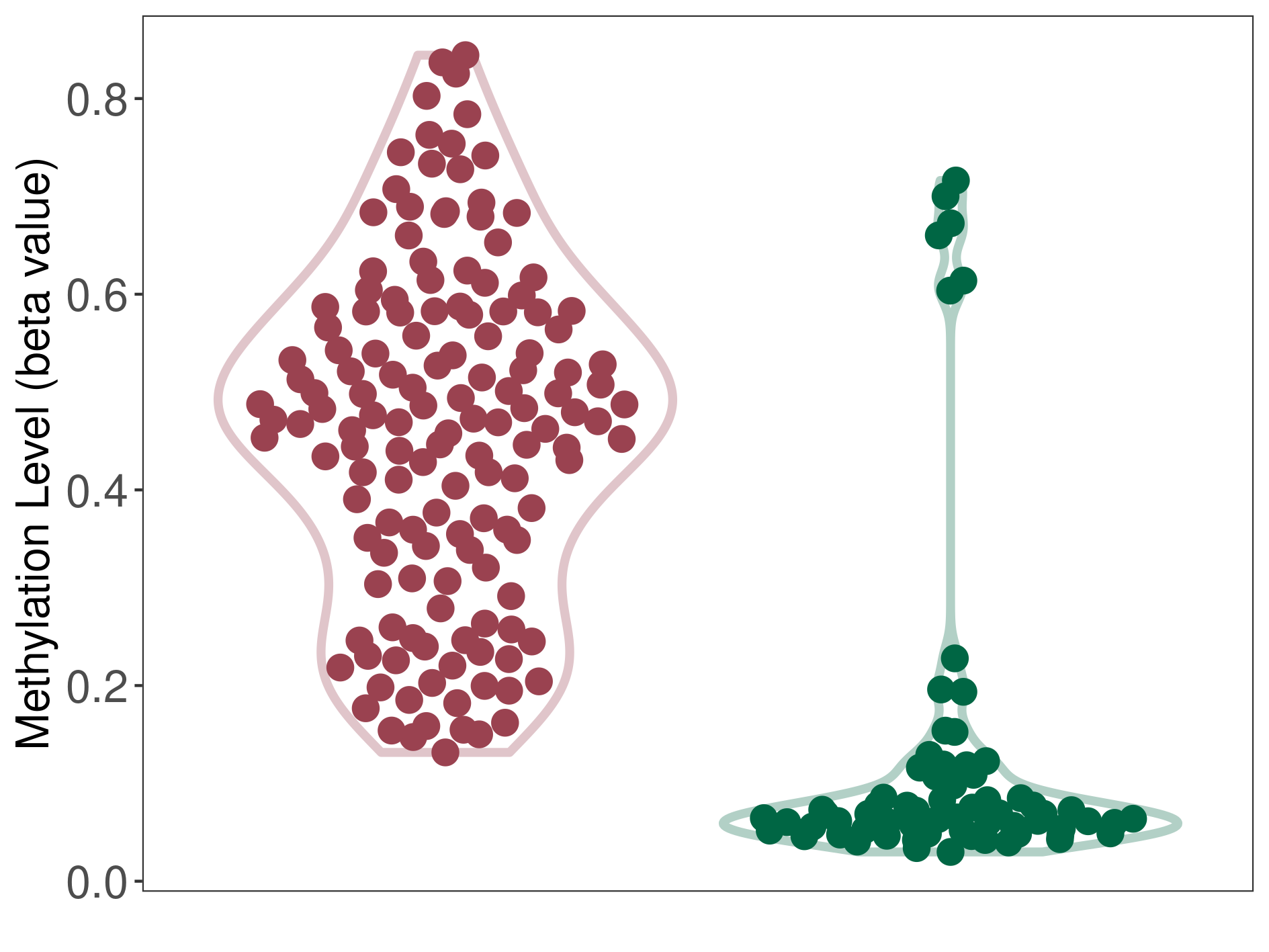
Epigenetic Phenomenon 2 |
Moderate hypermethylation of SLCO4C1 in prostate cancer than that in healthy individual | ||||
Studied Phenotype |
Prostate cancer [ICD-11:2C82] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 3.84E-29; Fold-change: 0.221026205; Z-score: 10.80695502 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
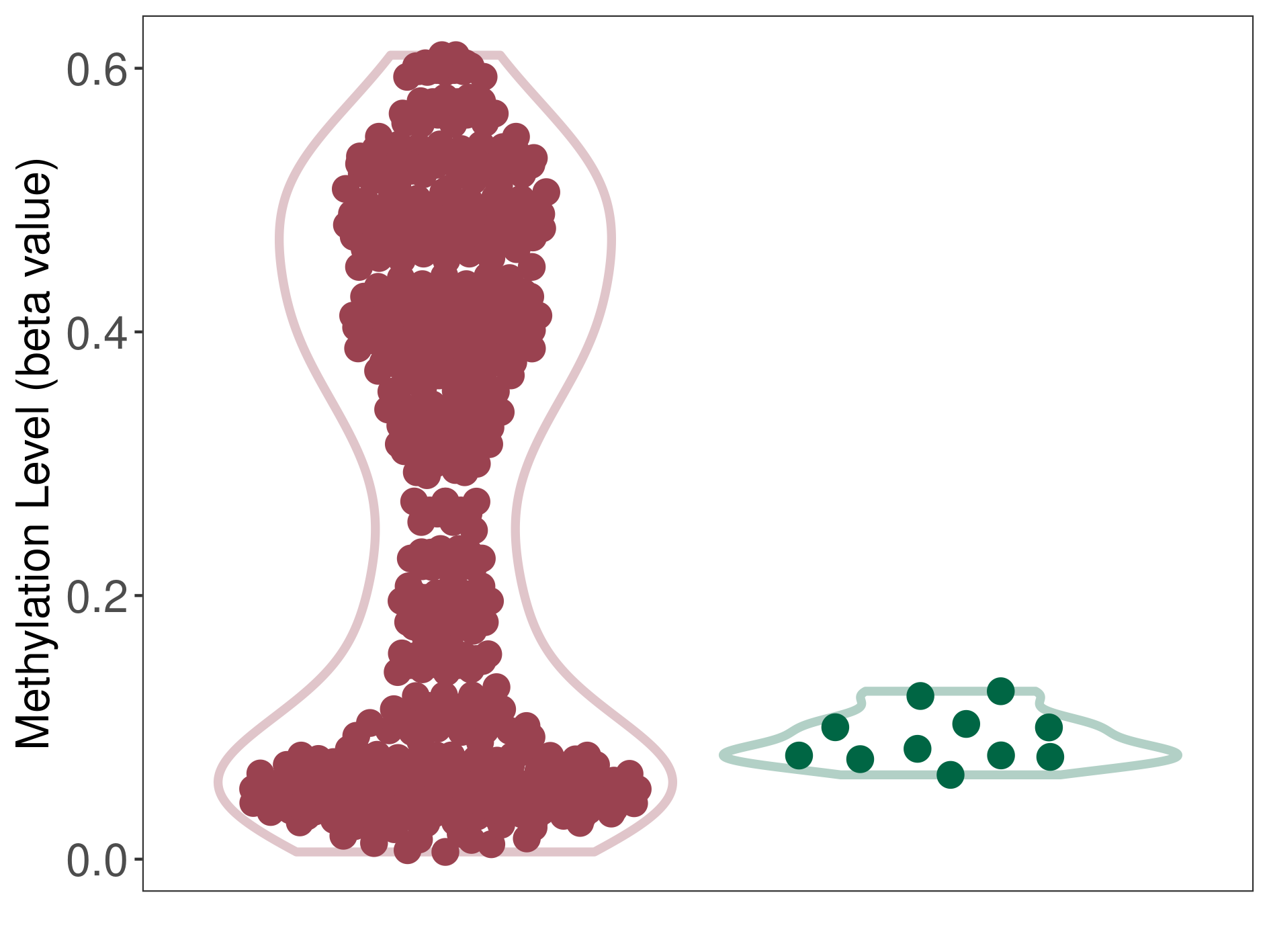
|
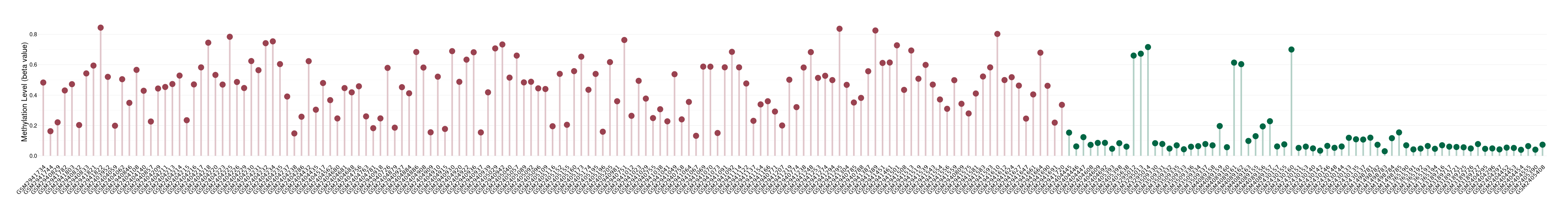
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Lung adenocarcinoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLCO4C1 in lung adenocarcinoma | [ 11 ] | |||
|
Location |
TSS200 (cg11009817) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.28E+00 | Statistic Test | p-value: 1.02E-02; Z-score: -1.76E+00 | ||
|
Methylation in Case |
9.22E-02 (Median) | Methylation in Control | 1.18E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Systemic lupus erythematosus |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLCO4C1 in systemic lupus erythematosus | [ 12 ] | |||
|
Location |
TSS200 (cg19788741) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.05E+00 | Statistic Test | p-value: 3.82E-03; Z-score: 1.68E-01 | ||
|
Methylation in Case |
2.67E-02 (Median) | Methylation in Control | 2.54E-02 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus [ ICD-11: 4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLCO4C1 in systemic lupus erythematosus | [ 12 ] | |||
|
Location |
TSS200 (cg06480736) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.04E+00 | Statistic Test | p-value: 5.84E-03; Z-score: 1.68E-01 | ||
|
Methylation in Case |
5.73E-02 (Median) | Methylation in Control | 5.52E-02 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus [ ICD-11: 4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of SLCO4C1 in systemic lupus erythematosus | [ 12 ] | |||
|
Location |
3'UTR (cg00630480) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.00E+00 | Statistic Test | p-value: 2.28E-02; Z-score: -5.57E-02 | ||
|
Methylation in Case |
8.32E-01 (Median) | Methylation in Control | 8.35E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus [ ICD-11: 4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Atypical teratoid rhabdoid tumor |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLCO4C1 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
1stExon (cg00056676) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -2.70E+00 | Statistic Test | p-value: 3.86E-42; Z-score: -6.50E+00 | ||
|
Methylation in Case |
2.99E-01 (Median) | Methylation in Control | 8.07E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLCO4C1 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
3'UTR (cg00630480) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.14E+00 | Statistic Test | p-value: 1.15E-15; Z-score: -2.99E+00 | ||
|
Methylation in Case |
7.78E-01 (Median) | Methylation in Control | 8.89E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor [ ICD-11: 2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Panic disorder |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of SLCO4C1 in panic disorder | [ 14 ] | |||
|
Location |
Body (cg23318786) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -9.92E-01 | Statistic Test | p-value: 2.58E-02; Z-score: -2.31E-01 | ||
|
Methylation in Case |
-5.48E+00 (Median) | Methylation in Control | -5.43E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder [ ICD-11: 6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of SLCO4C1 in panic disorder | [ 14 ] | |||
|
Location |
3'UTR (cg00630480) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.68E+00 | Statistic Test | p-value: 2.54E-06; Z-score: -8.11E-01 | ||
|
Methylation in Case |
5.29E-01 (Median) | Methylation in Control | 8.88E-01 (Median) | ||
|
Studied Phenotype |
Panic disorder [ ICD-11: 6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Glioblastoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypermethylation of SLCO4C1 in glioblastoma than that in healthy individual | ||||
Studied Phenotype |
Glioblastoma [ICD-11:2A00.00] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 2.03E-09; Fold-change: 0.481928989; Z-score: 2.094793725 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
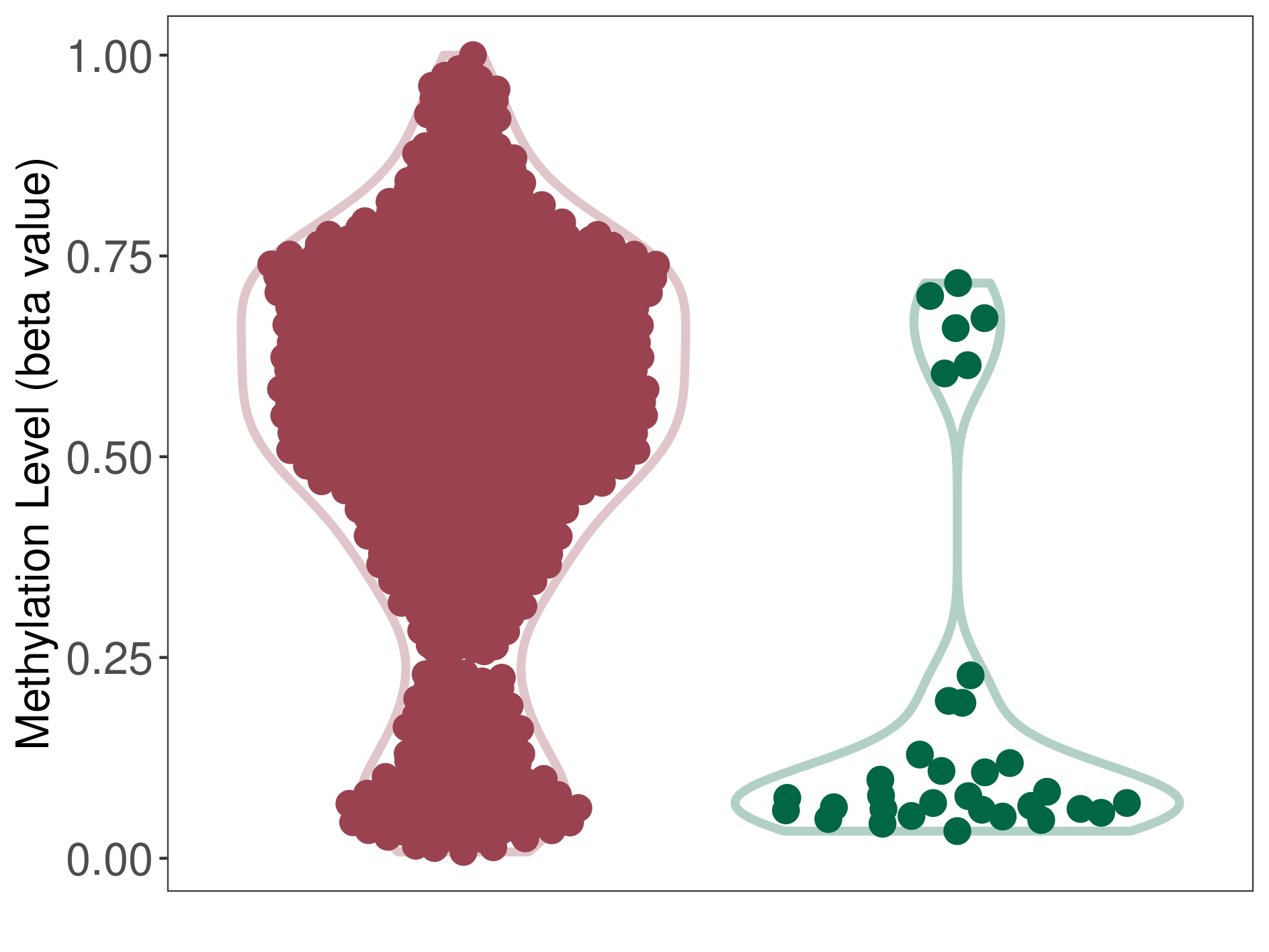
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Lymphoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypermethylation of SLCO4C1 in lymphoma than that in healthy individual | ||||
Studied Phenotype |
Lymphoma [ICD-11:2B30] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 4.10E-12; Fold-change: 0.469252292; Z-score: 2.714278546 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
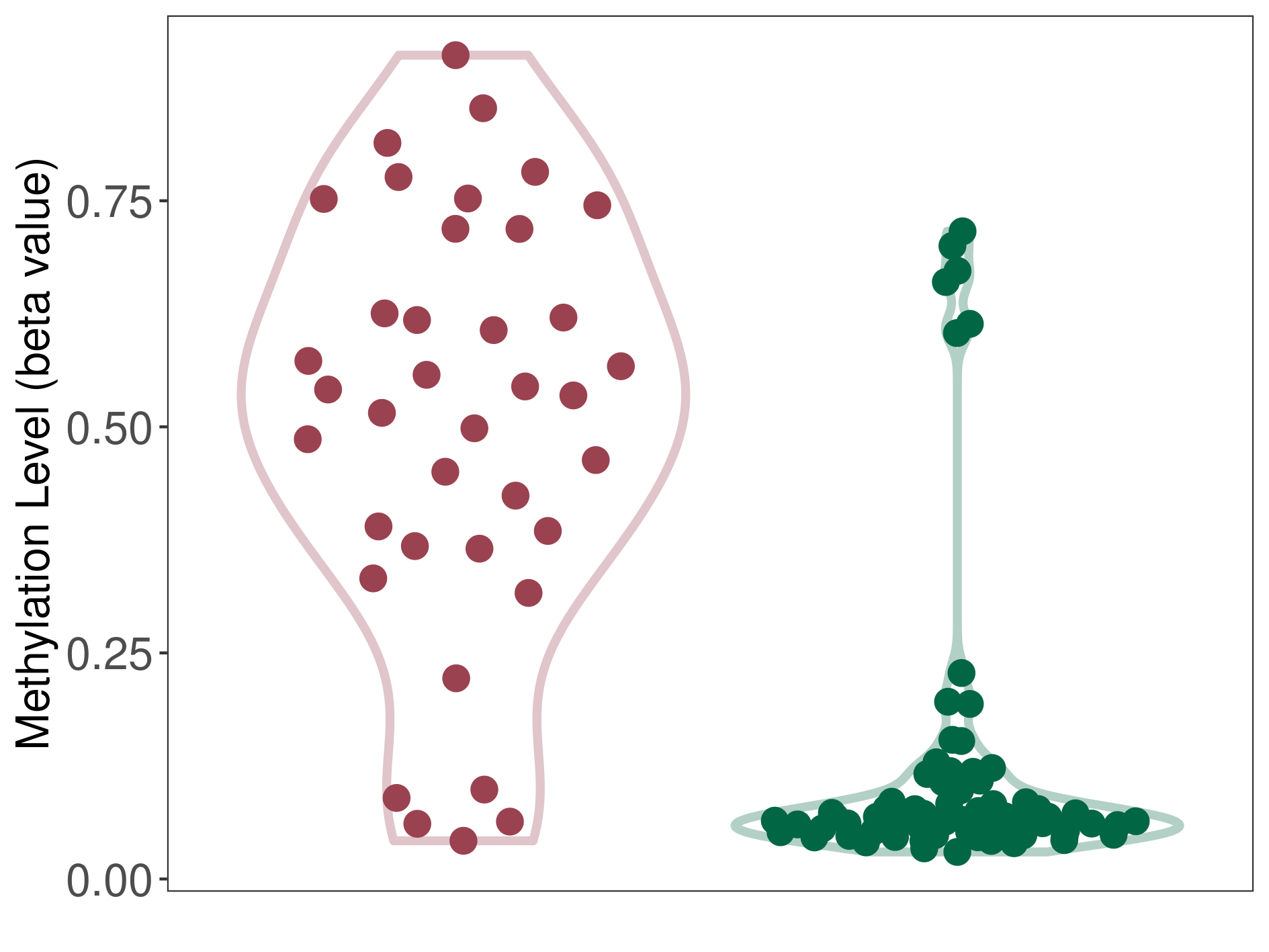
|
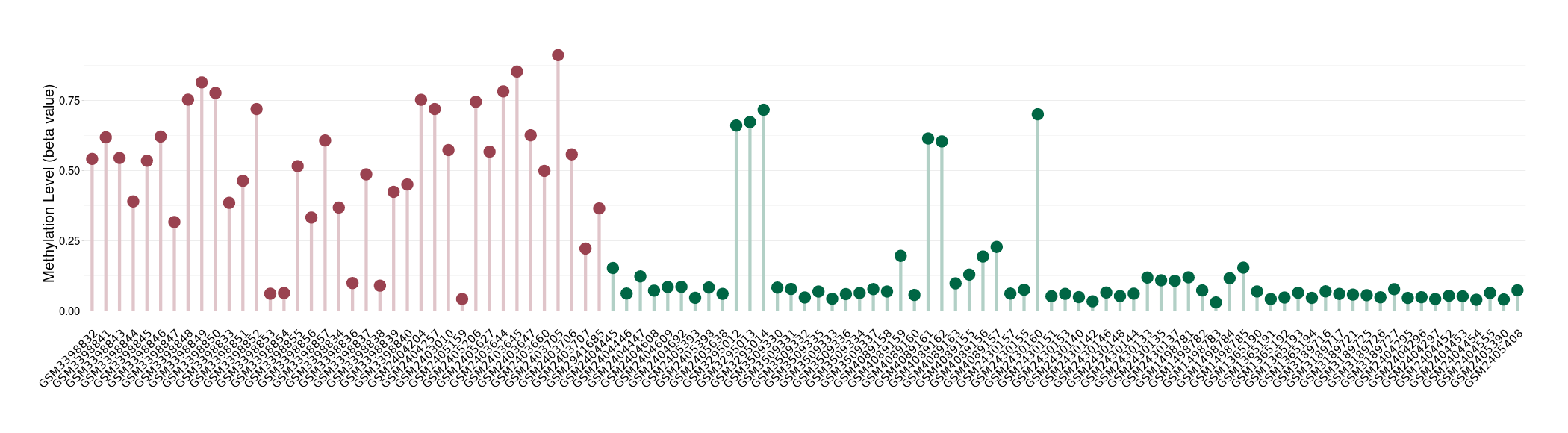 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
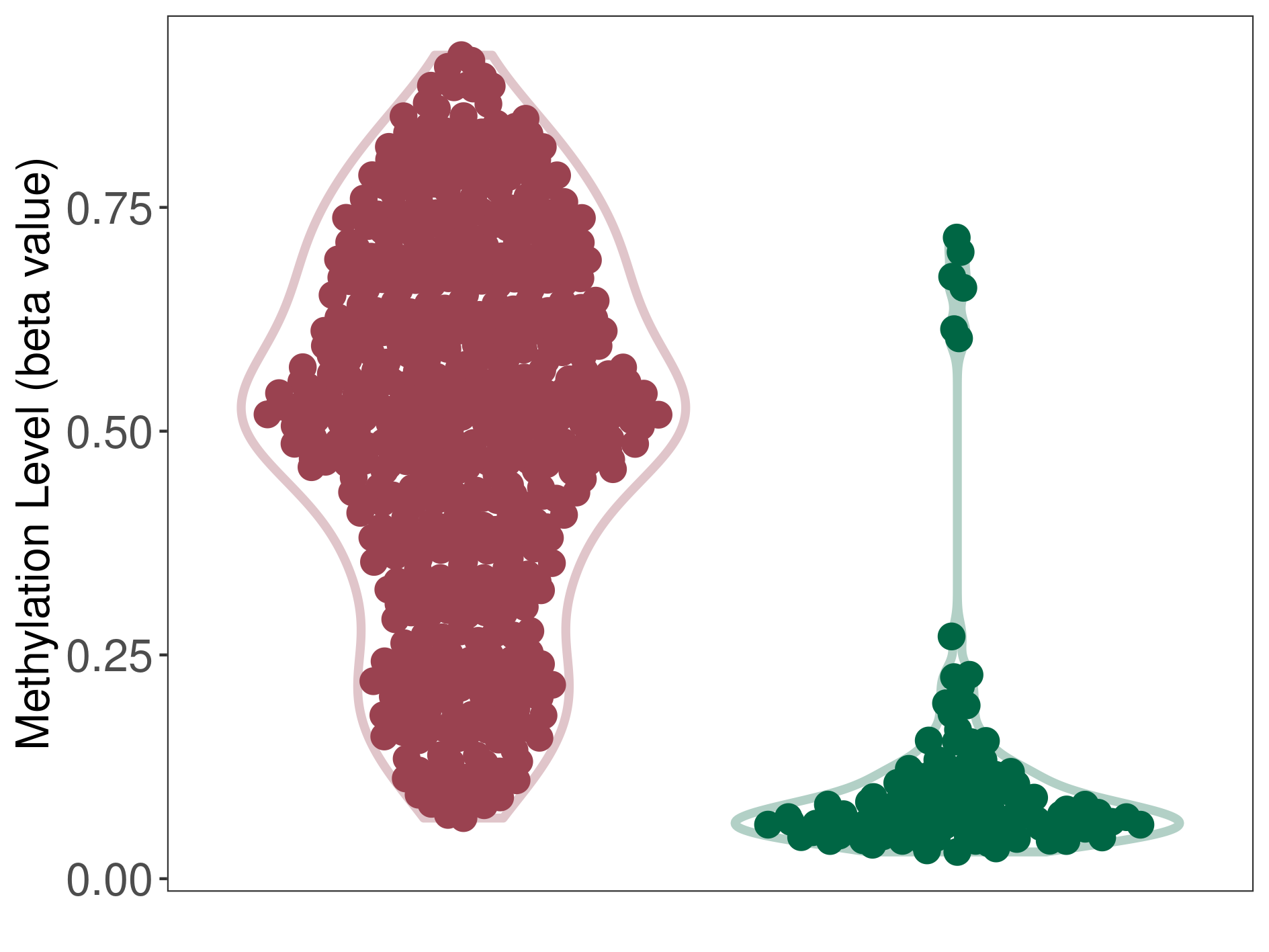
Malignant astrocytoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypermethylation of SLCO4C1 in malignant astrocytoma than that in healthy individual | ||||
Studied Phenotype |
Malignant astrocytoma [ICD-11:2A00.12] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 1.53E-64; Fold-change: 0.448187519; Z-score: 3.221077222 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
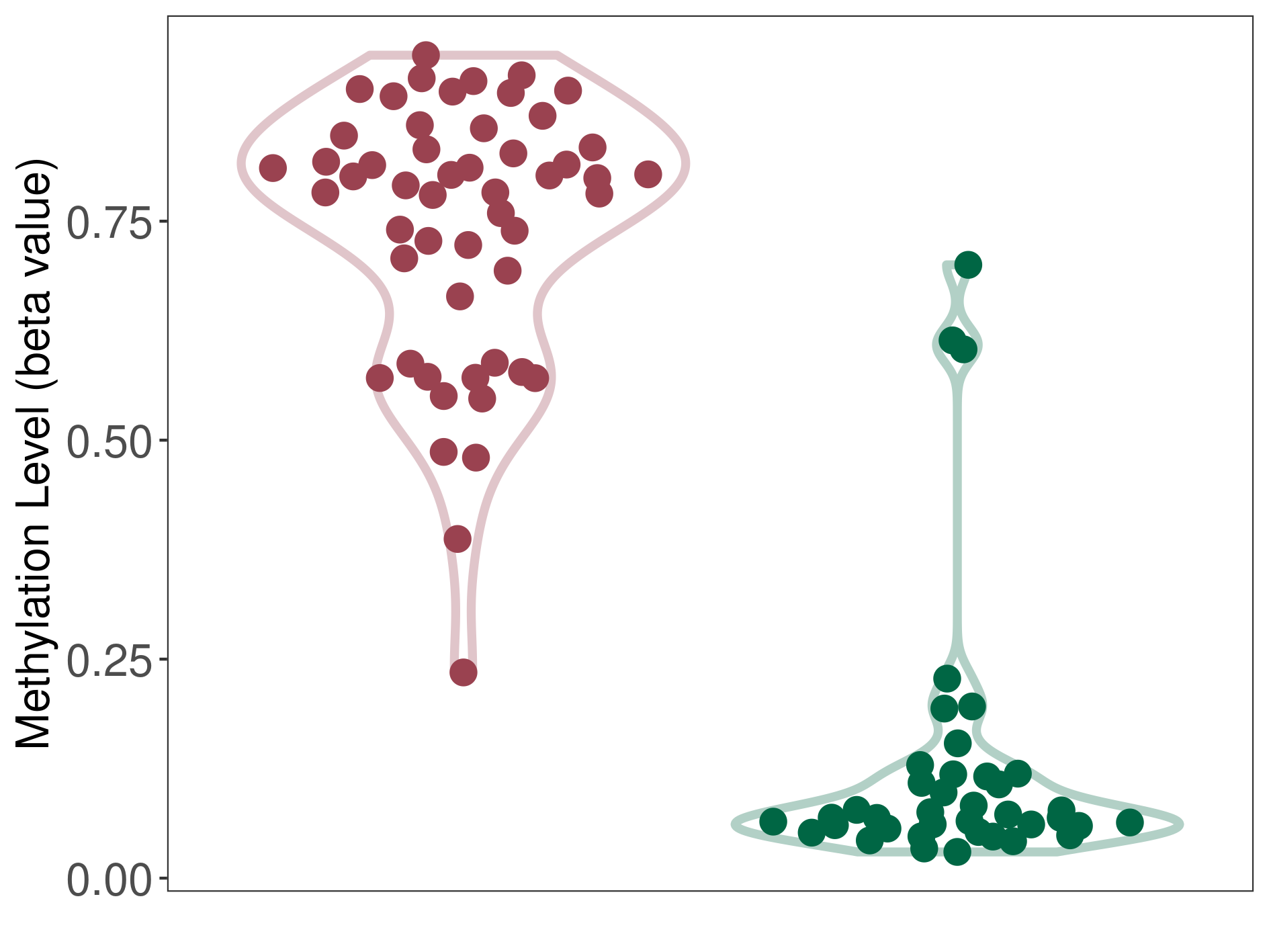
Multilayered rosettes embryonal tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypermethylation of SLCO4C1 in multilayered rosettes embryonal tumour than that in healthy individual | ||||
Studied Phenotype |
Multilayered rosettes embryonal tumour [ICD-11:2A00.1] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 4.00E-31; Fold-change: 0.725989969; Z-score: 4.631156229 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
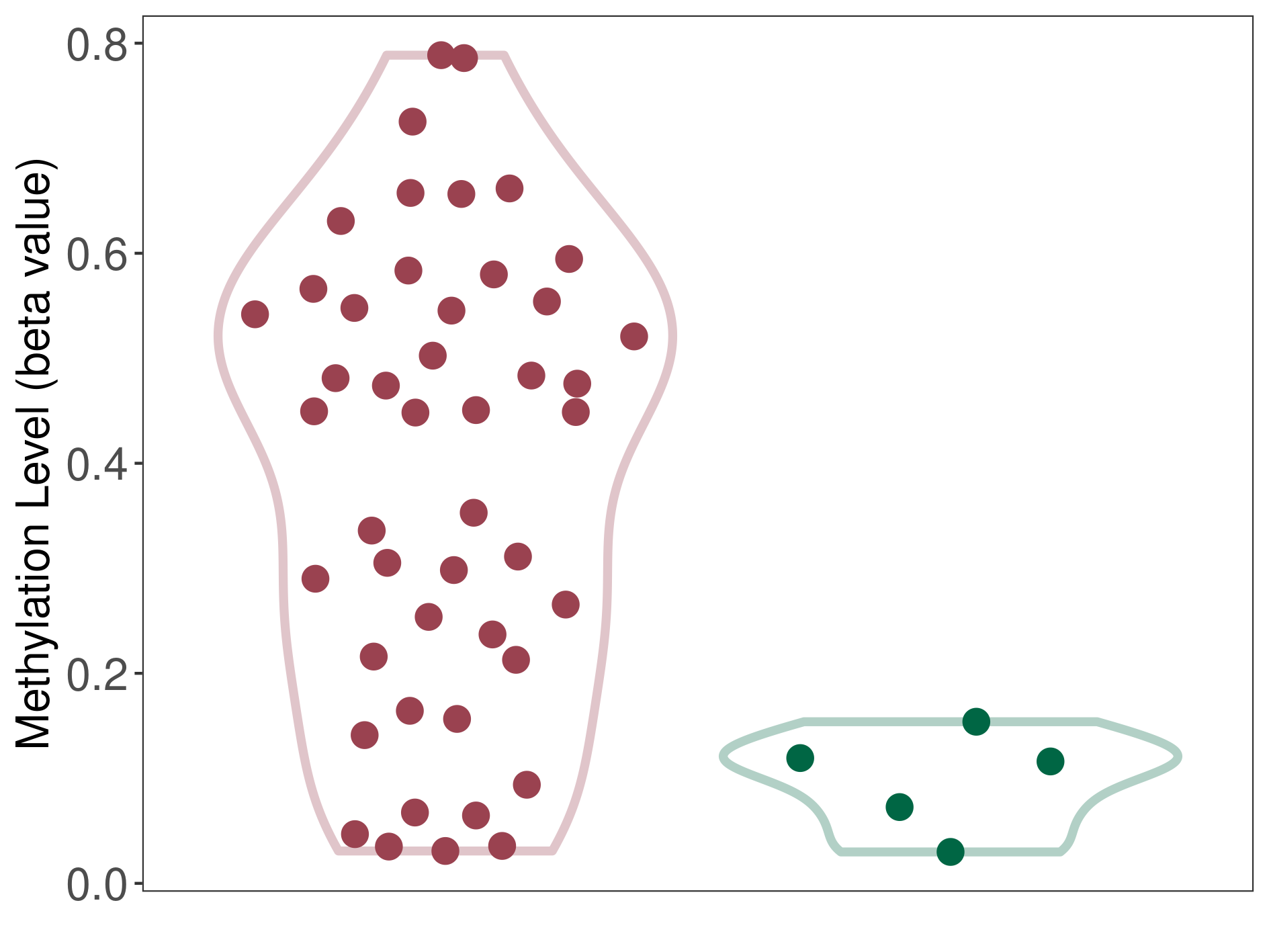
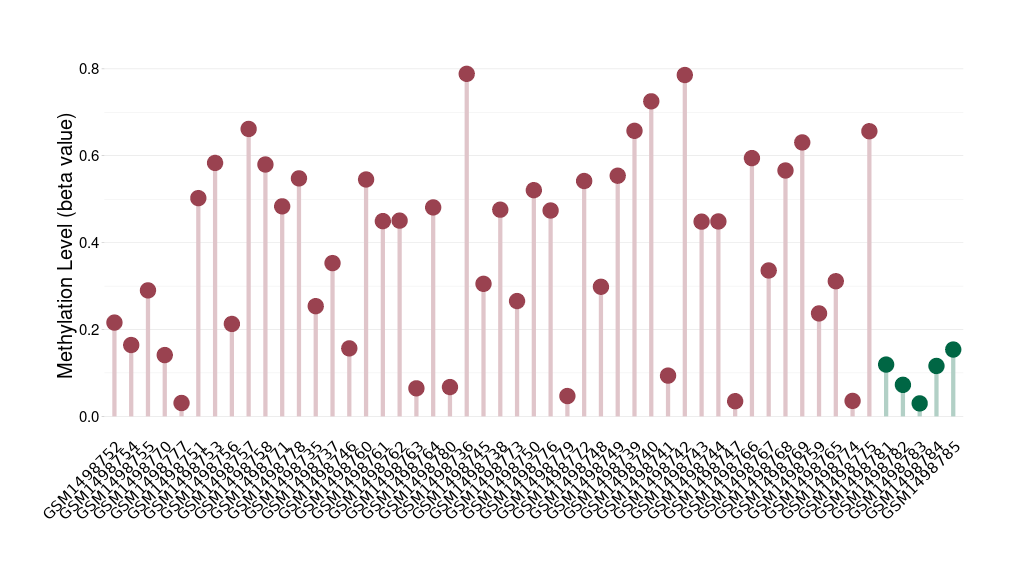
Oligodendroglial tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypermethylation of SLCO4C1 in oligodendroglial tumour than that in healthy individual | ||||
Studied Phenotype |
Oligodendroglial tumour [ICD-11:2A00.Y1] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 2.04E-08; Fold-change: 0.333051983; Z-score: 6.95185281 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Oligodendroglioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypermethylation of SLCO4C1 in oligodendroglioma than that in healthy individual | ||||
Studied Phenotype |
Oligodendroglioma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 6.00E-25; Fold-change: 0.402987977; Z-score: 2.33098834 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Prostate cancer metastasis |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
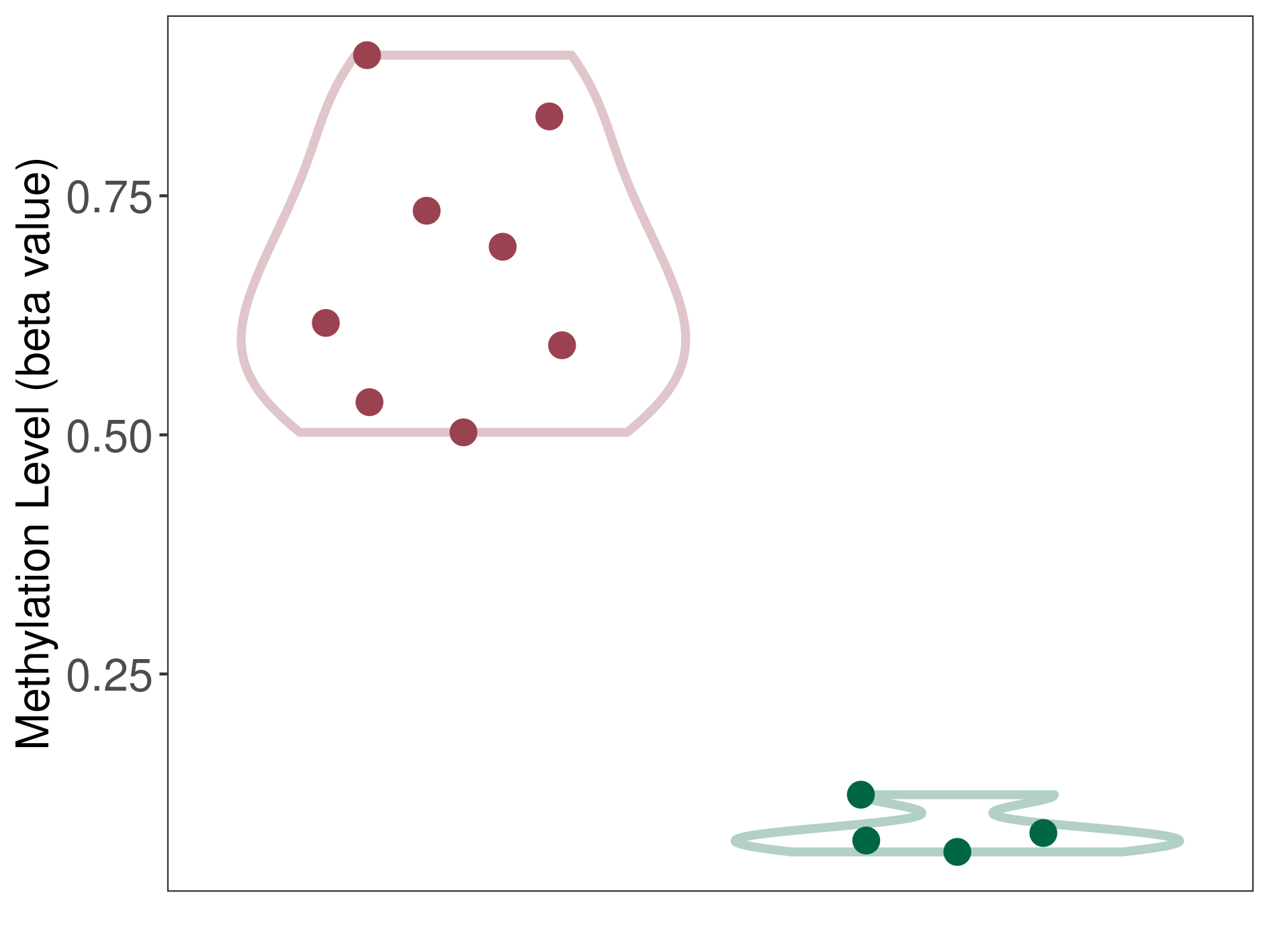
Epigenetic Phenomenon 1 |
Significant hypermethylation of SLCO4C1 in prostate cancer metastasis than that in healthy individual | ||||
Studied Phenotype |
Prostate cancer metastasis [ICD-11:2.00E+06] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 3.28E-06; Fold-change: 0.577116201; Z-score: 22.24530136 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
microRNA |
|||||
|
Unclear Phenotype |
25 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
miR-518f directly targets SLCO4C1 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Microarray | ||
|
Related Molecular Changes |
Down regulation of SLCO4C1 | Experiment Method | qRT-PCR | ||
|
miRNA Stemloop ID |
miR-518f | miRNA Mature ID | miR-518f-3p | ||
|
miRNA Sequence |
GAAAGCGCUUCUCUUUAGAGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
SLCO4C1 the predicted target of miR 518f-3p was determined in cell lysates of PBMCs infected with HIV-1 or HIV-2. | ||||
|
Epigenetic Phenomenon 2 |
miR-1185 directly targets SLCO4C1 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-1185 | miRNA Mature ID | miR-1185-5p | ||
|
miRNA Sequence |
AGAGGAUACCCUUUGUAUGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 3 |
miR-1224 directly targets SLCO4C1 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-1224 | miRNA Mature ID | miR-1224-5p | ||
|
miRNA Sequence |
GUGAGGACUCGGGAGGUGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 4 |
miR-124 directly targets SLCO4C1 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-124 | miRNA Mature ID | miR-124-5p | ||
|
miRNA Sequence |
CGUGUUCACAGCGGACCUUGAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon 5 |
miR-1264 directly targets SLCO4C1 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-1264 | miRNA Mature ID | miR-1264 | ||
|
miRNA Sequence |
CAAGUCUUAUUUGAGCACCUGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 6 |
miR-148b directly targets SLCO4C1 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Microarray | ||
|
miRNA Stemloop ID |
miR-148b | miRNA Mature ID | miR-148b-3p | ||
|
miRNA Sequence |
UCAGUGCAUCACAGAACUUUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human cervical cancer cell line (Hela) | ||||
|
Epigenetic Phenomenon 7 |
miR-2115 directly targets SLCO4C1 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-2115 | miRNA Mature ID | miR-2115-5p | ||
|
miRNA Sequence |
AGCUUCCAUGACUCCUGAUGGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 8 |
miR-335 directly targets SLCO4C1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Microarray | ||
|
miRNA Stemloop ID |
miR-335 | miRNA Mature ID | miR-335-5p | ||
|
miRNA Sequence |
UCAAGAGCAAUAACGAAAAAUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 9 |
miR-3605 directly targets SLCO4C1 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3605 | miRNA Mature ID | miR-3605-5p | ||
|
miRNA Sequence |
UGAGGAUGGAUAGCAAGGAAGCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 10 |
miR-3679 directly targets SLCO4C1 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3679 | miRNA Mature ID | miR-3679-5p | ||
|
miRNA Sequence |
UGAGGAUAUGGCAGGGAAGGGGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 11 |
miR-3692 directly targets SLCO4C1 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-3692 | miRNA Mature ID | miR-3692-3p | ||
|
miRNA Sequence |
GUUCCACACUGACACUGCAGAAGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 12 |
miR-3915 directly targets SLCO4C1 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3915 | miRNA Mature ID | miR-3915 | ||
|
miRNA Sequence |
UUGAGGAAAAGAUGGUCUUAUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 13 |
miR-3938 directly targets SLCO4C1 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-3938 | miRNA Mature ID | miR-3938 | ||
|
miRNA Sequence |
AAUUCCCUUGUAGAUAACCCGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 14 |
miR-4455 directly targets SLCO4C1 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4455 | miRNA Mature ID | miR-4455 | ||
|
miRNA Sequence |
AGGGUGUGUGUGUUUUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon 15 |
miR-4689 directly targets SLCO4C1 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4689 | miRNA Mature ID | miR-4689 | ||
|
miRNA Sequence |
UUGAGGAGACAUGGUGGGGGCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 16 |
miR-516a directly targets SLCO4C1 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-516a | miRNA Mature ID | miR-516a-3p | ||
|
miRNA Sequence |
UGCUUCCUUUCAGAGGGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 17 |
miR-516b directly targets SLCO4C1 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-516b | miRNA Mature ID | miR-516b-3p | ||
|
miRNA Sequence |
UGCUUCCUUUCAGAGGGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 18 |
miR-574 directly targets SLCO4C1 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-574 | miRNA Mature ID | miR-574-5p | ||
|
miRNA Sequence |
UGAGUGUGUGUGUGUGAGUGUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Left ventricular cardiac tissues of human | ||||
|
Epigenetic Phenomenon 19 |
miR-578 directly targets SLCO4C1 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-578 | miRNA Mature ID | miR-578 | ||
|
miRNA Sequence |
CUUCUUGUGCUCUAGGAUUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 20 |
miR-615 directly targets SLCO4C1 | [ 22 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | CLASH | ||
|
miRNA Stemloop ID |
miR-615 | miRNA Mature ID | miR-615-3p | ||
|
miRNA Sequence |
UCCGAGCCUGGGUCUCCCUCUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon 21 |
miR-670 directly targets SLCO4C1 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-670 | miRNA Mature ID | miR-670-5p | ||
|
miRNA Sequence |
GUCCCUGAGUGUAUGUGGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 22 |
miR-6752 directly targets SLCO4C1 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6752 | miRNA Mature ID | miR-6752-3p | ||
|
miRNA Sequence |
UCCCUGCCCCCAUACUCCCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 23 |
miR-676 directly targets SLCO4C1 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-676 | miRNA Mature ID | miR-676-5p | ||
|
miRNA Sequence |
UCUUCAACCUCAGGACUUGCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 24 |
miR-6858 directly targets SLCO4C1 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6858 | miRNA Mature ID | miR-6858-5p | ||
|
miRNA Sequence |
GUGAGGAGGGGCUGGCAGGGAC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon 25 |
miR-7162 directly targets SLCO4C1 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-7162 | miRNA Mature ID | miR-7162-5p | ||
|
miRNA Sequence |
UGCUUCCUUUCUCAGCUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.
