Detail Information of Epigenetic Regulations
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0523 Transporter Info | ||||
| Gene Name | CACNG2 | ||||
| Transporter Name | Voltage-dependent calcium channel gamma-2 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| Epigenetic Regulations of This DT (EGR) | |||||
|---|---|---|---|---|---|
|
Methylation |
|||||
|
Prostate cancer |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of CACNG2 in prostate cancer | [ 1 ] | |||
|
Location |
5'UTR (cg04011829) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.69E+00 | Statistic Test | p-value: 2.50E-03; Z-score: 5.22E+00 | ||
|
Methylation in Case |
7.11E-01 (Median) | Methylation in Control | 4.22E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of CACNG2 in prostate cancer | [ 1 ] | |||
|
Location |
Body (cg08181642) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.16E+00 | Statistic Test | p-value: 1.75E-02; Z-score: 9.39E+00 | ||
|
Methylation in Case |
8.37E-01 (Median) | Methylation in Control | 7.24E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of CACNG2 in prostate cancer | [ 1 ] | |||
|
Location |
Body (cg00161124) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.25E+00 | Statistic Test | p-value: 4.81E-02; Z-score: 2.19E+00 | ||
|
Methylation in Case |
7.29E-01 (Median) | Methylation in Control | 5.81E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer [ ICD-11: 2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Bladder cancer |
10 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of CACNG2 in bladder cancer | [ 2 ] | |||
|
Location |
TSS1500 (cg12451153) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.25E+00 | Statistic Test | p-value: 1.79E-02; Z-score: -2.10E+00 | ||
|
Methylation in Case |
8.30E-02 (Median) | Methylation in Control | 1.03E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of CACNG2 in bladder cancer | [ 2 ] | |||
|
Location |
TSS200 (cg17580331) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -2.04E+00 | Statistic Test | p-value: 2.56E-03; Z-score: -3.79E+00 | ||
|
Methylation in Case |
1.49E-01 (Median) | Methylation in Control | 3.05E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of CACNG2 in bladder cancer | [ 2 ] | |||
|
Location |
Body (cg15606745) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.32E+00 | Statistic Test | p-value: 3.80E-08; Z-score: -1.29E+01 | ||
|
Methylation in Case |
4.59E-01 (Median) | Methylation in Control | 6.06E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of CACNG2 in bladder cancer | [ 2 ] | |||
|
Location |
Body (cg18666630) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.95E+00 | Statistic Test | p-value: 6.09E-08; Z-score: -7.04E+00 | ||
|
Methylation in Case |
3.08E-01 (Median) | Methylation in Control | 6.00E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of CACNG2 in bladder cancer | [ 2 ] | |||
|
Location |
Body (cg18984186) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.29E+00 | Statistic Test | p-value: 4.83E-07; Z-score: -9.58E+00 | ||
|
Methylation in Case |
5.59E-01 (Median) | Methylation in Control | 7.23E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of CACNG2 in bladder cancer | [ 2 ] | |||
|
Location |
Body (cg04350355) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.23E+00 | Statistic Test | p-value: 2.39E-06; Z-score: -9.69E+00 | ||
|
Methylation in Case |
6.47E-01 (Median) | Methylation in Control | 7.97E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of CACNG2 in bladder cancer | [ 2 ] | |||
|
Location |
Body (cg06943853) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.19E+00 | Statistic Test | p-value: 5.21E-06; Z-score: -6.97E+00 | ||
|
Methylation in Case |
6.80E-01 (Median) | Methylation in Control | 8.10E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of CACNG2 in bladder cancer | [ 2 ] | |||
|
Location |
Body (cg04933168) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.24E+00 | Statistic Test | p-value: 3.01E-05; Z-score: -3.28E+00 | ||
|
Methylation in Case |
5.78E-01 (Median) | Methylation in Control | 7.17E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of CACNG2 in bladder cancer | [ 2 ] | |||
|
Location |
Body (cg04186249) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.25E+00 | Statistic Test | p-value: 1.07E-04; Z-score: -4.73E+00 | ||
|
Methylation in Case |
5.94E-01 (Median) | Methylation in Control | 7.45E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 10 |
Methylation of CACNG2 in bladder cancer | [ 2 ] | |||
|
Location |
Body (cg17806661) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.39E+00 | Statistic Test | p-value: 2.43E-04; Z-score: -3.51E+00 | ||
|
Methylation in Case |
2.52E-01 (Median) | Methylation in Control | 3.49E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer [ ICD-11: 2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Breast cancer |
13 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of CACNG2 in breast cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg01151699) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.15E+00 | Statistic Test | p-value: 9.39E-04; Z-score: 3.85E-01 | ||
|
Methylation in Case |
4.47E-02 (Median) | Methylation in Control | 3.90E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of CACNG2 in breast cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg20648240) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.21E+00 | Statistic Test | p-value: 4.15E-03; Z-score: 8.06E-01 | ||
|
Methylation in Case |
4.92E-02 (Median) | Methylation in Control | 4.05E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of CACNG2 in breast cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg12451153) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.20E+00 | Statistic Test | p-value: 7.01E-03; Z-score: 4.85E-01 | ||
|
Methylation in Case |
8.61E-02 (Median) | Methylation in Control | 7.18E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of CACNG2 in breast cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg08242574) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.16E+00 | Statistic Test | p-value: 1.76E-02; Z-score: 4.10E-01 | ||
|
Methylation in Case |
5.70E-02 (Median) | Methylation in Control | 4.93E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of CACNG2 in breast cancer | [ 3 ] | |||
|
Location |
TSS200 (cg24290616) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.33E+00 | Statistic Test | p-value: 1.16E-05; Z-score: 1.09E+00 | ||
|
Methylation in Case |
8.31E-02 (Median) | Methylation in Control | 6.24E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of CACNG2 in breast cancer | [ 3 ] | |||
|
Location |
Body (cg13270055) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.60E+00 | Statistic Test | p-value: 1.18E-24; Z-score: 3.55E+00 | ||
|
Methylation in Case |
5.90E-01 (Median) | Methylation in Control | 3.70E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of CACNG2 in breast cancer | [ 3 ] | |||
|
Location |
Body (cg04186249) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.21E+00 | Statistic Test | p-value: 9.40E-12; Z-score: -2.56E+00 | ||
|
Methylation in Case |
5.71E-01 (Median) | Methylation in Control | 6.94E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of CACNG2 in breast cancer | [ 3 ] | |||
|
Location |
Body (cg18666630) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.19E+00 | Statistic Test | p-value: 3.72E-08; Z-score: -1.65E+00 | ||
|
Methylation in Case |
4.58E-01 (Median) | Methylation in Control | 5.46E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of CACNG2 in breast cancer | [ 3 ] | |||
|
Location |
Body (cg15606745) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.03E+00 | Statistic Test | p-value: 1.57E-04; Z-score: -6.35E-01 | ||
|
Methylation in Case |
5.63E-01 (Median) | Methylation in Control | 5.81E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 10 |
Methylation of CACNG2 in breast cancer | [ 3 ] | |||
|
Location |
Body (cg04933168) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.08E+00 | Statistic Test | p-value: 3.59E-03; Z-score: 1.00E+00 | ||
|
Methylation in Case |
6.90E-01 (Median) | Methylation in Control | 6.36E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 11 |
Methylation of CACNG2 in breast cancer | [ 3 ] | |||
|
Location |
Body (cg04350355) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.07E+00 | Statistic Test | p-value: 7.75E-03; Z-score: 9.87E-01 | ||
|
Methylation in Case |
7.13E-01 (Median) | Methylation in Control | 6.64E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 12 |
Methylation of CACNG2 in breast cancer | [ 3 ] | |||
|
Location |
Body (cg22496723) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.03E+00 | Statistic Test | p-value: 2.58E-02; Z-score: 5.10E-01 | ||
|
Methylation in Case |
8.30E-01 (Median) | Methylation in Control | 8.05E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 13 |
Methylation of CACNG2 in breast cancer | [ 3 ] | |||
|
Location |
Body (cg18984186) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.06E+00 | Statistic Test | p-value: 4.21E-02; Z-score: -7.65E-01 | ||
|
Methylation in Case |
6.62E-01 (Median) | Methylation in Control | 7.02E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer [ ICD-11: 2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colorectal cancer |
15 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of CACNG2 in colorectal cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg01151699) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.07E+00 | Statistic Test | p-value: 1.72E-05; Z-score: -3.10E-01 | ||
|
Methylation in Case |
8.97E-02 (Median) | Methylation in Control | 9.57E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of CACNG2 in colorectal cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg12451153) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.07E+00 | Statistic Test | p-value: 6.77E-04; Z-score: 2.62E-01 | ||
|
Methylation in Case |
1.51E-01 (Median) | Methylation in Control | 1.41E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of CACNG2 in colorectal cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg08242574) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.04E+00 | Statistic Test | p-value: 2.83E-03; Z-score: 1.60E-01 | ||
|
Methylation in Case |
1.48E-01 (Median) | Methylation in Control | 1.43E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of CACNG2 in colorectal cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg20648240) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.03E+00 | Statistic Test | p-value: 6.80E-03; Z-score: -1.10E-01 | ||
|
Methylation in Case |
5.71E-02 (Median) | Methylation in Control | 5.90E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of CACNG2 in colorectal cancer | [ 4 ] | |||
|
Location |
TSS200 (cg04191954) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.03E+00 | Statistic Test | p-value: 1.89E-05; Z-score: 1.75E-01 | ||
|
Methylation in Case |
2.17E-01 (Median) | Methylation in Control | 2.10E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of CACNG2 in colorectal cancer | [ 4 ] | |||
|
Location |
TSS200 (cg17580331) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.19E+00 | Statistic Test | p-value: 2.24E-03; Z-score: 1.68E+00 | ||
|
Methylation in Case |
5.69E-01 (Median) | Methylation in Control | 4.78E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of CACNG2 in colorectal cancer | [ 4 ] | |||
|
Location |
TSS200 (cg24290616) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.06E+00 | Statistic Test | p-value: 4.25E-03; Z-score: -2.34E-01 | ||
|
Methylation in Case |
2.35E-01 (Median) | Methylation in Control | 2.49E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of CACNG2 in colorectal cancer | [ 4 ] | |||
|
Location |
Body (cg17806661) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.41E+00 | Statistic Test | p-value: 3.25E-14; Z-score: -3.40E+00 | ||
|
Methylation in Case |
4.79E-01 (Median) | Methylation in Control | 6.78E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of CACNG2 in colorectal cancer | [ 4 ] | |||
|
Location |
Body (cg15606745) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.24E+00 | Statistic Test | p-value: 2.01E-13; Z-score: -3.10E+00 | ||
|
Methylation in Case |
5.33E-01 (Median) | Methylation in Control | 6.64E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 10 |
Methylation of CACNG2 in colorectal cancer | [ 4 ] | |||
|
Location |
Body (cg04186249) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.18E+00 | Statistic Test | p-value: 3.18E-13; Z-score: -5.02E+00 | ||
|
Methylation in Case |
7.27E-01 (Median) | Methylation in Control | 8.61E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 11 |
Methylation of CACNG2 in colorectal cancer | [ 4 ] | |||
|
Location |
Body (cg06943853) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.13E+00 | Statistic Test | p-value: 5.49E-10; Z-score: -3.52E+00 | ||
|
Methylation in Case |
7.45E-01 (Median) | Methylation in Control | 8.42E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 12 |
Methylation of CACNG2 in colorectal cancer | [ 4 ] | |||
|
Location |
Body (cg18984186) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.06E+00 | Statistic Test | p-value: 5.11E-09; Z-score: -2.86E+00 | ||
|
Methylation in Case |
7.94E-01 (Median) | Methylation in Control | 8.44E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 13 |
Methylation of CACNG2 in colorectal cancer | [ 4 ] | |||
|
Location |
Body (cg04350355) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.07E+00 | Statistic Test | p-value: 5.06E-07; Z-score: -3.33E+00 | ||
|
Methylation in Case |
8.20E-01 (Median) | Methylation in Control | 8.80E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 14 |
Methylation of CACNG2 in colorectal cancer | [ 4 ] | |||
|
Location |
Body (cg04933168) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.03E+00 | Statistic Test | p-value: 1.42E-03; Z-score: -8.53E-01 | ||
|
Methylation in Case |
8.30E-01 (Median) | Methylation in Control | 8.55E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 15 |
Methylation of CACNG2 in colorectal cancer | [ 4 ] | |||
|
Location |
Body (cg18666630) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.18E+00 | Statistic Test | p-value: 1.82E-03; Z-score: -8.33E-01 | ||
|
Methylation in Case |
4.35E-01 (Median) | Methylation in Control | 5.14E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer [ ICD-11: 2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Hepatocellular carcinoma |
14 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of CACNG2 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg05159370) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.45E+00 | Statistic Test | p-value: 2.80E-11; Z-score: -3.23E+00 | ||
|
Methylation in Case |
4.09E-01 (Median) | Methylation in Control | 5.93E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of CACNG2 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg12451153) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.13E+00 | Statistic Test | p-value: 2.11E-05; Z-score: 4.92E-01 | ||
|
Methylation in Case |
1.59E-01 (Median) | Methylation in Control | 1.41E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of CACNG2 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg01151699) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.24E+00 | Statistic Test | p-value: 2.38E-05; Z-score: 1.05E+00 | ||
|
Methylation in Case |
8.01E-02 (Median) | Methylation in Control | 6.49E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of CACNG2 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg20648240) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.08E+00 | Statistic Test | p-value: 5.67E-03; Z-score: 3.08E-01 | ||
|
Methylation in Case |
6.61E-02 (Median) | Methylation in Control | 6.11E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of CACNG2 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg08242574) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.10E+00 | Statistic Test | p-value: 5.90E-03; Z-score: 2.22E-01 | ||
|
Methylation in Case |
9.60E-02 (Median) | Methylation in Control | 8.76E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of CACNG2 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS200 (cg25212131) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.24E+00 | Statistic Test | p-value: 4.15E-18; Z-score: 2.62E+00 | ||
|
Methylation in Case |
8.21E-01 (Median) | Methylation in Control | 6.65E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of CACNG2 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS200 (cg04191954) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.03E+00 | Statistic Test | p-value: 5.31E-03; Z-score: -1.69E-01 | ||
|
Methylation in Case |
1.04E-01 (Median) | Methylation in Control | 1.08E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of CACNG2 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
Body (cg23368787) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.38E+00 | Statistic Test | p-value: 2.74E-15; Z-score: 2.94E+00 | ||
|
Methylation in Case |
6.12E-01 (Median) | Methylation in Control | 4.43E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of CACNG2 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
Body (cg12848639) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.36E+00 | Statistic Test | p-value: 2.08E-13; Z-score: -5.62E+00 | ||
|
Methylation in Case |
5.06E-01 (Median) | Methylation in Control | 6.86E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 10 |
Methylation of CACNG2 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
Body (cg12072789) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.30E+00 | Statistic Test | p-value: 2.37E-10; Z-score: -2.07E+00 | ||
|
Methylation in Case |
4.77E-01 (Median) | Methylation in Control | 6.19E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 11 |
Methylation of CACNG2 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
Body (cg04437482) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.24E+00 | Statistic Test | p-value: 9.24E-10; Z-score: -2.25E+00 | ||
|
Methylation in Case |
5.57E-01 (Median) | Methylation in Control | 6.89E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 12 |
Methylation of CACNG2 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
Body (cg18984186) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.05E+00 | Statistic Test | p-value: 3.13E-05; Z-score: -1.13E+00 | ||
|
Methylation in Case |
6.75E-01 (Median) | Methylation in Control | 7.12E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 13 |
Methylation of CACNG2 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
Body (cg22496723) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.09E+00 | Statistic Test | p-value: 2.82E-03; Z-score: -1.14E+00 | ||
|
Methylation in Case |
7.20E-01 (Median) | Methylation in Control | 7.87E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 14 |
Methylation of CACNG2 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
Body (cg13270055) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.11E+00 | Statistic Test | p-value: 5.54E-03; Z-score: -6.92E-01 | ||
|
Methylation in Case |
4.53E-01 (Median) | Methylation in Control | 5.03E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma [ ICD-11: 2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Lung adenocarcinoma |
6 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of CACNG2 in lung adenocarcinoma | [ 6 ] | |||
|
Location |
TSS1500 (cg08242574) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.34E+00 | Statistic Test | p-value: 4.26E-03; Z-score: 1.28E+00 | ||
|
Methylation in Case |
1.44E-01 (Median) | Methylation in Control | 1.07E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of CACNG2 in lung adenocarcinoma | [ 6 ] | |||
|
Location |
TSS1500 (cg01151699) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.44E+00 | Statistic Test | p-value: 3.15E-02; Z-score: 2.27E+00 | ||
|
Methylation in Case |
1.13E-01 (Median) | Methylation in Control | 7.85E-02 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of CACNG2 in lung adenocarcinoma | [ 6 ] | |||
|
Location |
Body (cg04933168) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.09E+00 | Statistic Test | p-value: 9.86E-03; Z-score: 1.33E+00 | ||
|
Methylation in Case |
7.91E-01 (Median) | Methylation in Control | 7.23E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of CACNG2 in lung adenocarcinoma | [ 6 ] | |||
|
Location |
Body (cg18666630) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.12E+00 | Statistic Test | p-value: 1.76E-02; Z-score: -2.01E+00 | ||
|
Methylation in Case |
5.57E-01 (Median) | Methylation in Control | 6.21E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of CACNG2 in lung adenocarcinoma | [ 6 ] | |||
|
Location |
Body (cg13270055) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.16E+00 | Statistic Test | p-value: 4.07E-02; Z-score: 9.07E-01 | ||
|
Methylation in Case |
7.18E-01 (Median) | Methylation in Control | 6.18E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of CACNG2 in lung adenocarcinoma | [ 6 ] | |||
|
Location |
Body (cg15606745) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.04E+00 | Statistic Test | p-value: 4.71E-02; Z-score: -1.54E+00 | ||
|
Methylation in Case |
5.73E-01 (Median) | Methylation in Control | 5.97E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma [ ICD-11: 2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Pancretic ductal adenocarcinoma |
10 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of CACNG2 in pancretic ductal adenocarcinoma | [ 7 ] | |||
|
Location |
TSS1500 (cg14918082) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.20E+00 | Statistic Test | p-value: 7.40E-04; Z-score: -1.08E+00 | ||
|
Methylation in Case |
5.87E-01 (Median) | Methylation in Control | 7.03E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of CACNG2 in pancretic ductal adenocarcinoma | [ 7 ] | |||
|
Location |
TSS1500 (cg10508416) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.06E+00 | Statistic Test | p-value: 1.52E-03; Z-score: -7.08E-01 | ||
|
Methylation in Case |
7.54E-01 (Median) | Methylation in Control | 8.02E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of CACNG2 in pancretic ductal adenocarcinoma | [ 7 ] | |||
|
Location |
TSS1500 (cg26904702) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.04E+00 | Statistic Test | p-value: 9.54E-03; Z-score: -6.14E-01 | ||
|
Methylation in Case |
7.18E-01 (Median) | Methylation in Control | 7.44E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of CACNG2 in pancretic ductal adenocarcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg15014549) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.10E+00 | Statistic Test | p-value: 2.88E-09; Z-score: 5.61E-01 | ||
|
Methylation in Case |
1.01E-01 (Median) | Methylation in Control | 9.19E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of CACNG2 in pancretic ductal adenocarcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg18771173) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.52E+00 | Statistic Test | p-value: 5.33E-05; Z-score: 6.09E-01 | ||
|
Methylation in Case |
1.22E-01 (Median) | Methylation in Control | 8.03E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of CACNG2 in pancretic ductal adenocarcinoma | [ 7 ] | |||
|
Location |
Body (cg01643580) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.21E+00 | Statistic Test | p-value: 1.67E-06; Z-score: 1.32E+00 | ||
|
Methylation in Case |
3.45E-01 (Median) | Methylation in Control | 2.86E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 7 |
Methylation of CACNG2 in pancretic ductal adenocarcinoma | [ 7 ] | |||
|
Location |
Body (cg06347454) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.08E+00 | Statistic Test | p-value: 5.57E-04; Z-score: 8.57E-01 | ||
|
Methylation in Case |
8.37E-01 (Median) | Methylation in Control | 7.75E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 8 |
Methylation of CACNG2 in pancretic ductal adenocarcinoma | [ 7 ] | |||
|
Location |
Body (cg09168805) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.05E+00 | Statistic Test | p-value: 1.30E-03; Z-score: 3.10E-01 | ||
|
Methylation in Case |
7.45E-01 (Median) | Methylation in Control | 7.11E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 9 |
Methylation of CACNG2 in pancretic ductal adenocarcinoma | [ 7 ] | |||
|
Location |
Body (cg00706648) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.03E+00 | Statistic Test | p-value: 5.20E-03; Z-score: 6.74E-01 | ||
|
Methylation in Case |
7.31E-01 (Median) | Methylation in Control | 7.08E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 10 |
Methylation of CACNG2 in pancretic ductal adenocarcinoma | [ 7 ] | |||
|
Location |
Body (cg24115232) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.19E+00 | Statistic Test | p-value: 1.95E-02; Z-score: 9.12E-01 | ||
|
Methylation in Case |
5.50E-01 (Median) | Methylation in Control | 4.63E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma [ ICD-11: 2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Papillary thyroid cancer |
6 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of CACNG2 in papillary thyroid cancer | [ 8 ] | |||
|
Location |
TSS1500 (cg12451153) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.01E+00 | Statistic Test | p-value: 2.17E-02; Z-score: 4.76E-02 | ||
|
Methylation in Case |
6.94E-02 (Median) | Methylation in Control | 6.85E-02 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of CACNG2 in papillary thyroid cancer | [ 8 ] | |||
|
Location |
Body (cg04186249) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.12E+00 | Statistic Test | p-value: 5.67E-11; Z-score: -1.83E+00 | ||
|
Methylation in Case |
7.32E-01 (Median) | Methylation in Control | 8.17E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of CACNG2 in papillary thyroid cancer | [ 8 ] | |||
|
Location |
Body (cg15606745) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.04E+00 | Statistic Test | p-value: 6.47E-06; Z-score: -1.12E+00 | ||
|
Methylation in Case |
5.58E-01 (Median) | Methylation in Control | 5.82E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of CACNG2 in papillary thyroid cancer | [ 8 ] | |||
|
Location |
Body (cg18666630) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.06E+00 | Statistic Test | p-value: 5.97E-04; Z-score: -6.14E-01 | ||
|
Methylation in Case |
6.73E-01 (Median) | Methylation in Control | 7.11E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 5 |
Methylation of CACNG2 in papillary thyroid cancer | [ 8 ] | |||
|
Location |
Body (cg04350355) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.02E+00 | Statistic Test | p-value: 9.57E-04; Z-score: -6.08E-01 | ||
|
Methylation in Case |
8.83E-01 (Median) | Methylation in Control | 9.03E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 6 |
Methylation of CACNG2 in papillary thyroid cancer | [ 8 ] | |||
|
Location |
Body (cg06943853) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.02E+00 | Statistic Test | p-value: 3.76E-03; Z-score: -5.51E-01 | ||
|
Methylation in Case |
7.74E-01 (Median) | Methylation in Control | 7.89E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer [ ICD-11: 2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Renal cell carcinoma |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of CACNG2 in clear cell renal cell carcinoma | [ 9 ] | |||
|
Location |
TSS200 (cg24290616) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.42E+00 | Statistic Test | p-value: 4.12E-03; Z-score: 8.82E-01 | ||
|
Methylation in Case |
3.30E-02 (Median) | Methylation in Control | 2.32E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of CACNG2 in clear cell renal cell carcinoma | [ 9 ] | |||
|
Location |
TSS200 (cg04191954) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.23E+00 | Statistic Test | p-value: 4.50E-03; Z-score: 7.44E-01 | ||
|
Methylation in Case |
4.27E-02 (Median) | Methylation in Control | 3.47E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of CACNG2 in clear cell renal cell carcinoma | [ 9 ] | |||
|
Location |
Body (cg13270055) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.22E+00 | Statistic Test | p-value: 4.55E-03; Z-score: 1.33E+00 | ||
|
Methylation in Case |
7.52E-01 (Median) | Methylation in Control | 6.15E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma [ ICD-11: 2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colon cancer |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of CACNG2 in colon adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg09150064) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.49E+00 | Statistic Test | p-value: 6.68E-04; Z-score: 2.47E+00 | ||
|
Methylation in Case |
3.68E-01 (Median) | Methylation in Control | 2.46E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of CACNG2 in colon adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg02857074) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.26E+00 | Statistic Test | p-value: 7.03E-04; Z-score: -1.11E+00 | ||
|
Methylation in Case |
3.06E-01 (Median) | Methylation in Control | 3.86E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of CACNG2 in colon adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg20050113) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -1.27E+00 | Statistic Test | p-value: 7.22E-04; Z-score: -1.75E+00 | ||
|
Methylation in Case |
3.12E-01 (Median) | Methylation in Control | 3.98E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer [ ICD-11: 2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Panic disorder |
4 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Methylation of CACNG2 in panic disorder | [ 11 ] | |||
|
Location |
Body (cg17806661) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: -9.04E-01 | Statistic Test | p-value: 1.75E-03; Z-score: -4.46E-01 | ||
|
Methylation in Case |
-1.74E+00 (Median) | Methylation in Control | -1.58E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder [ ICD-11: 6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 2 |
Methylation of CACNG2 in panic disorder | [ 11 ] | |||
|
Location |
Body (cg04186249) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.11E+00 | Statistic Test | p-value: 1.09E-02; Z-score: 4.76E-01 | ||
|
Methylation in Case |
2.20E+00 (Median) | Methylation in Control | 1.99E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder [ ICD-11: 6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 3 |
Methylation of CACNG2 in panic disorder | [ 11 ] | |||
|
Location |
Body (cg18666630) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.10E+00 | Statistic Test | p-value: 2.59E-02; Z-score: 1.68E-01 | ||
|
Methylation in Case |
7.49E-01 (Median) | Methylation in Control | 6.79E-01 (Median) | ||
|
Studied Phenotype |
Panic disorder [ ICD-11: 6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon 4 |
Methylation of CACNG2 in panic disorder | [ 11 ] | |||
|
Location |
Body (cg04350355) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change: 1.07E+00 | Statistic Test | p-value: 3.65E-02; Z-score: 4.12E-01 | ||
|
Methylation in Case |
3.21E+00 (Median) | Methylation in Control | 3.01E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder [ ICD-11: 6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
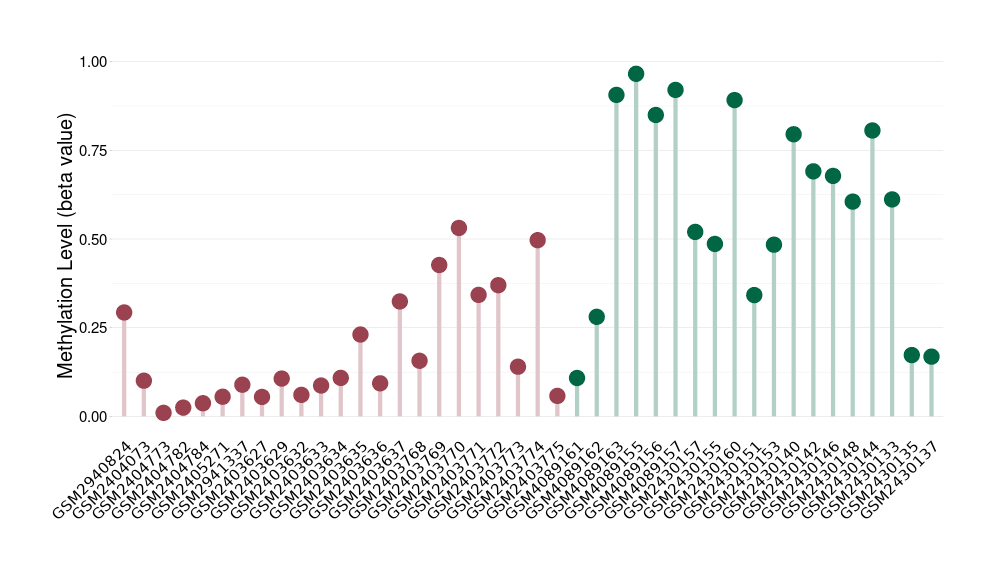
Chordoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Moderate hypermethylation of CACNG2 in chordoma than that in healthy individual | ||||
Studied Phenotype |
Chordoma [ICD-11:5A61.0] | ||||
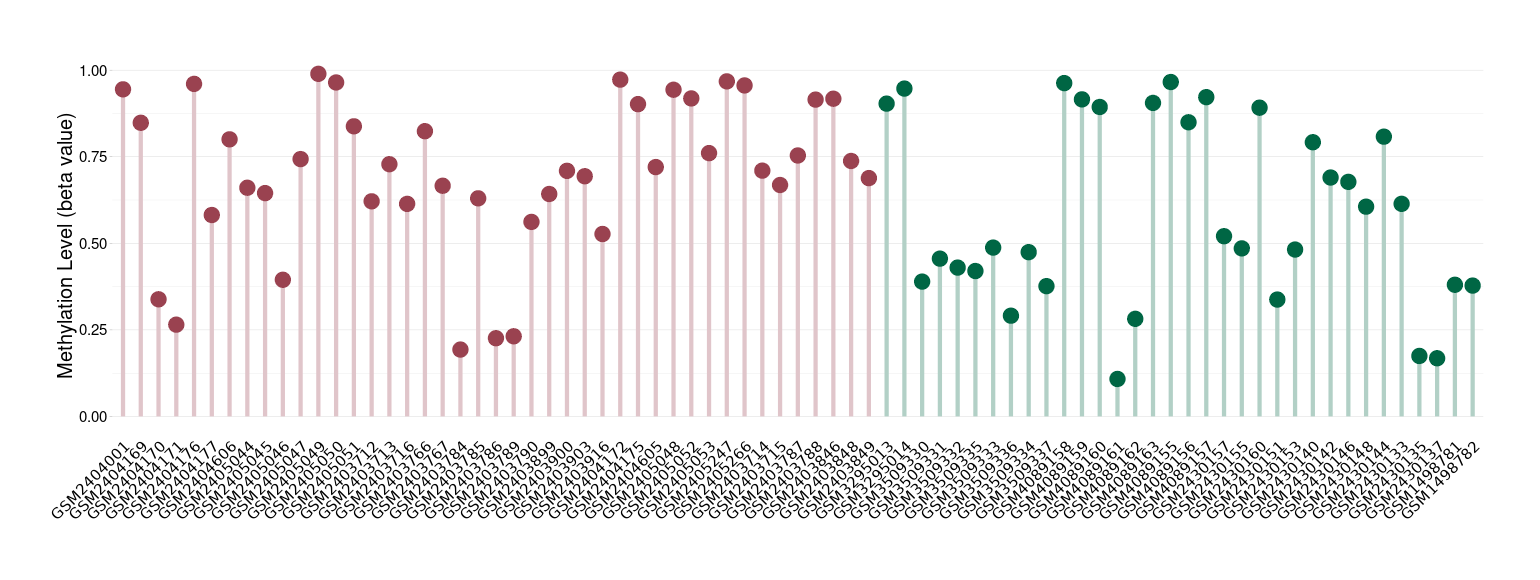
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 0.004919165; Fold-change: 0.243965097; Z-score: 1.167112221 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
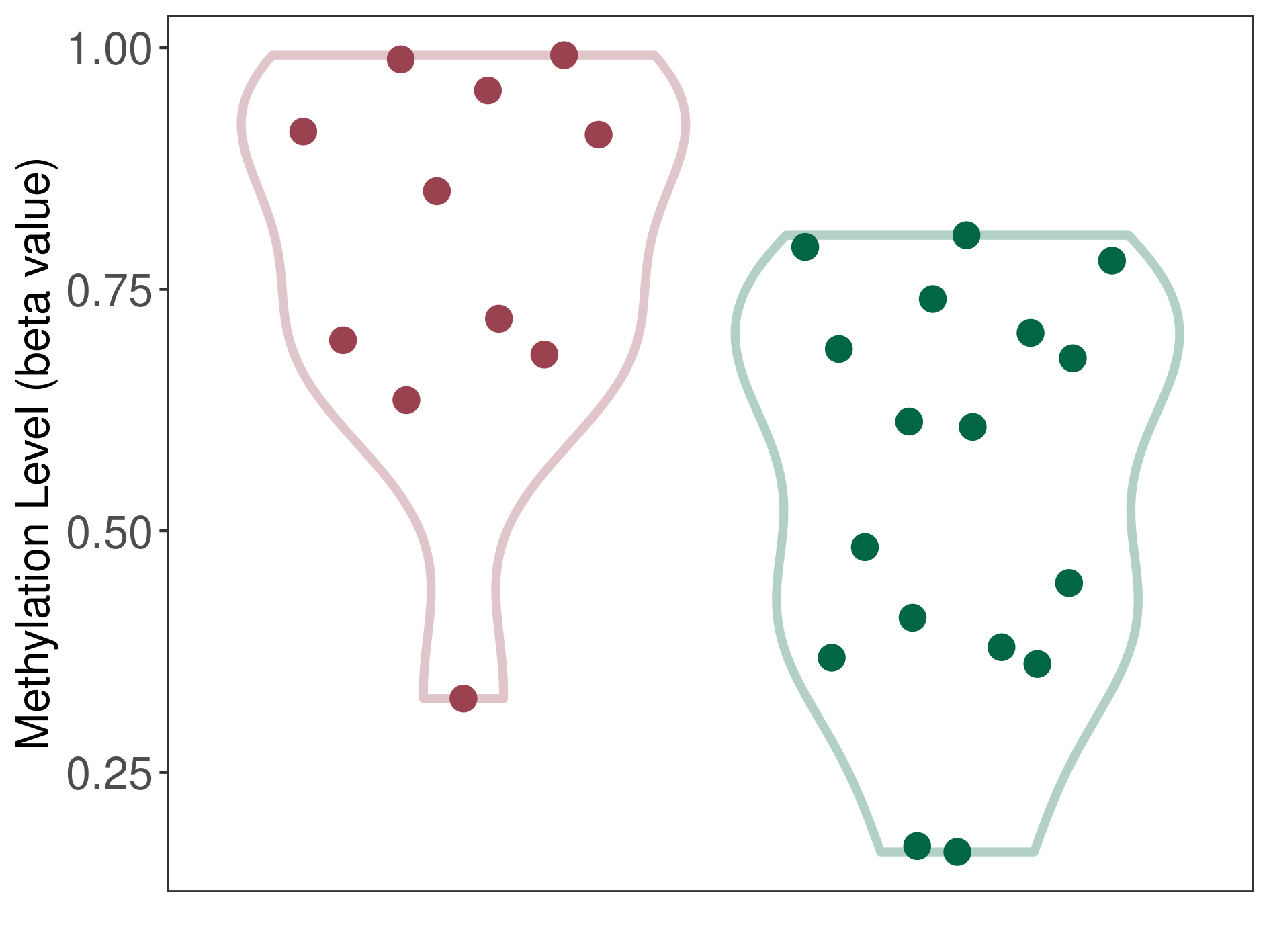
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Craniopharyngioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Moderate hypermethylation of CACNG2 in craniopharyngioma than that in healthy individual | ||||
Studied Phenotype |
Craniopharyngioma [ICD-11:2F9A] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 0.039252322; Fold-change: 0.216139585; Z-score: 0.8170911 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
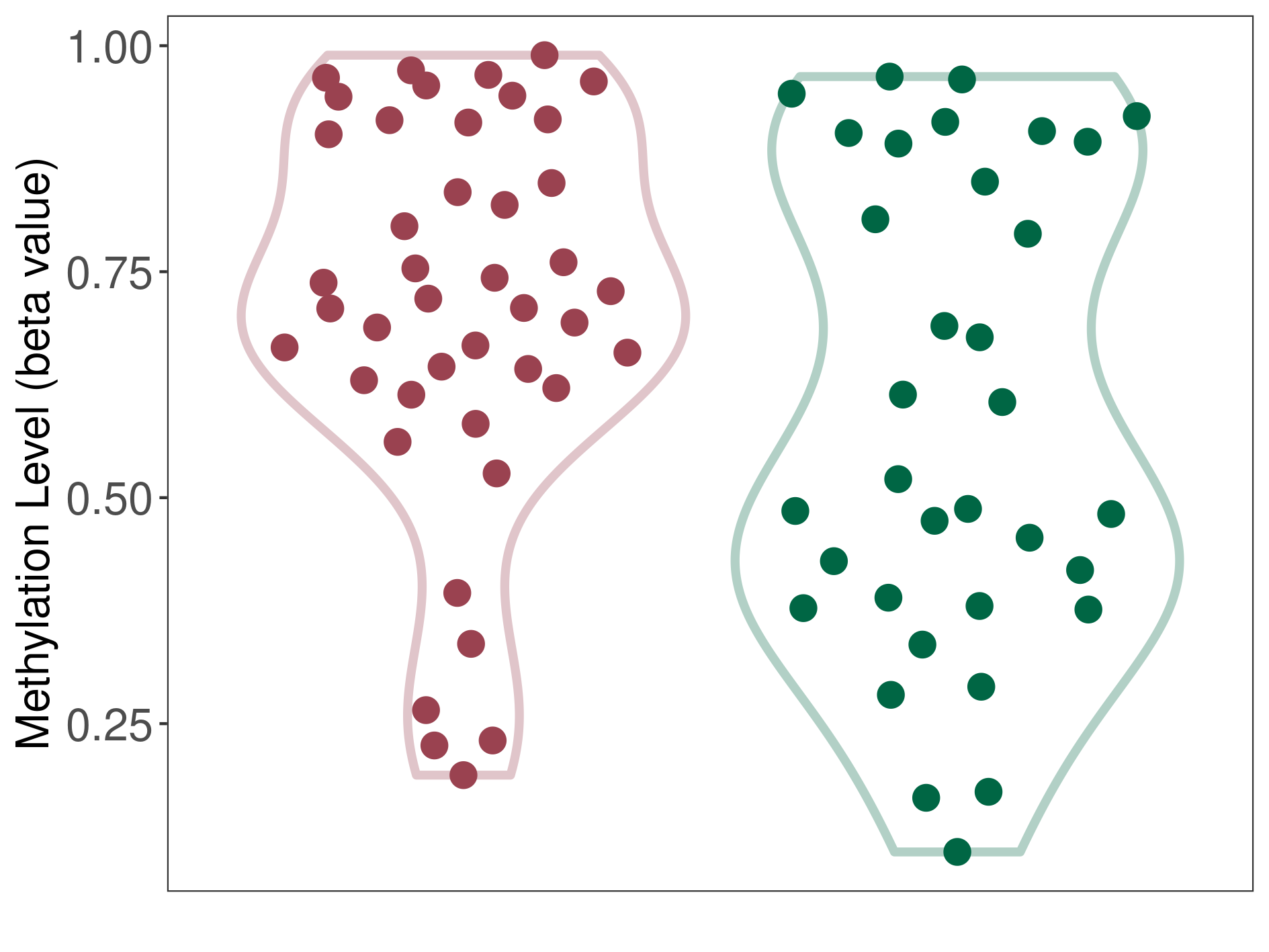
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
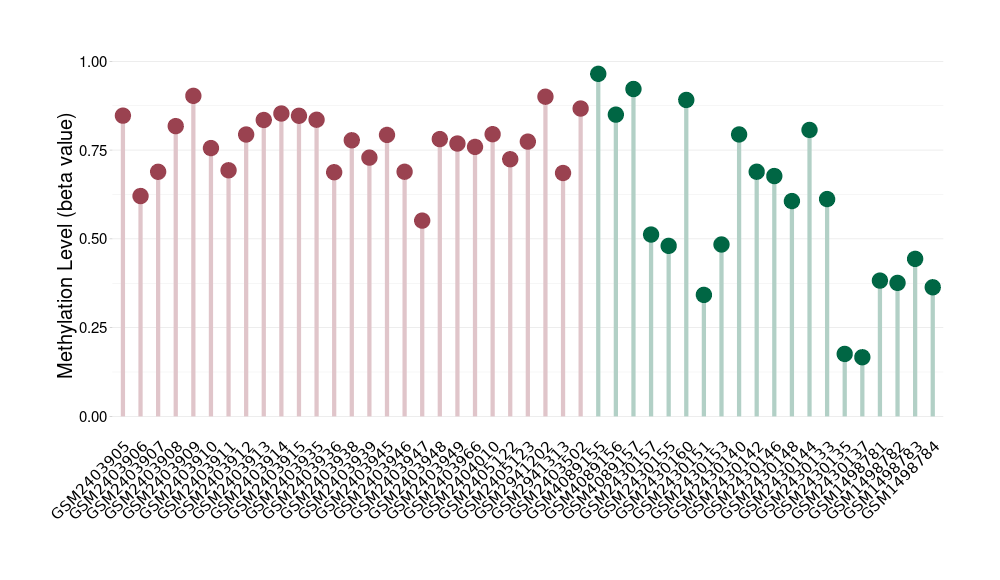
Hemangioblastoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Moderate hypermethylation of CACNG2 in hemangioblastoma than that in healthy individual | ||||
Studied Phenotype |
Hemangioblastoma [ICD-11:2F7C] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 0.002515146; Fold-change: 0.218404137; Z-score: 0.901451947 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
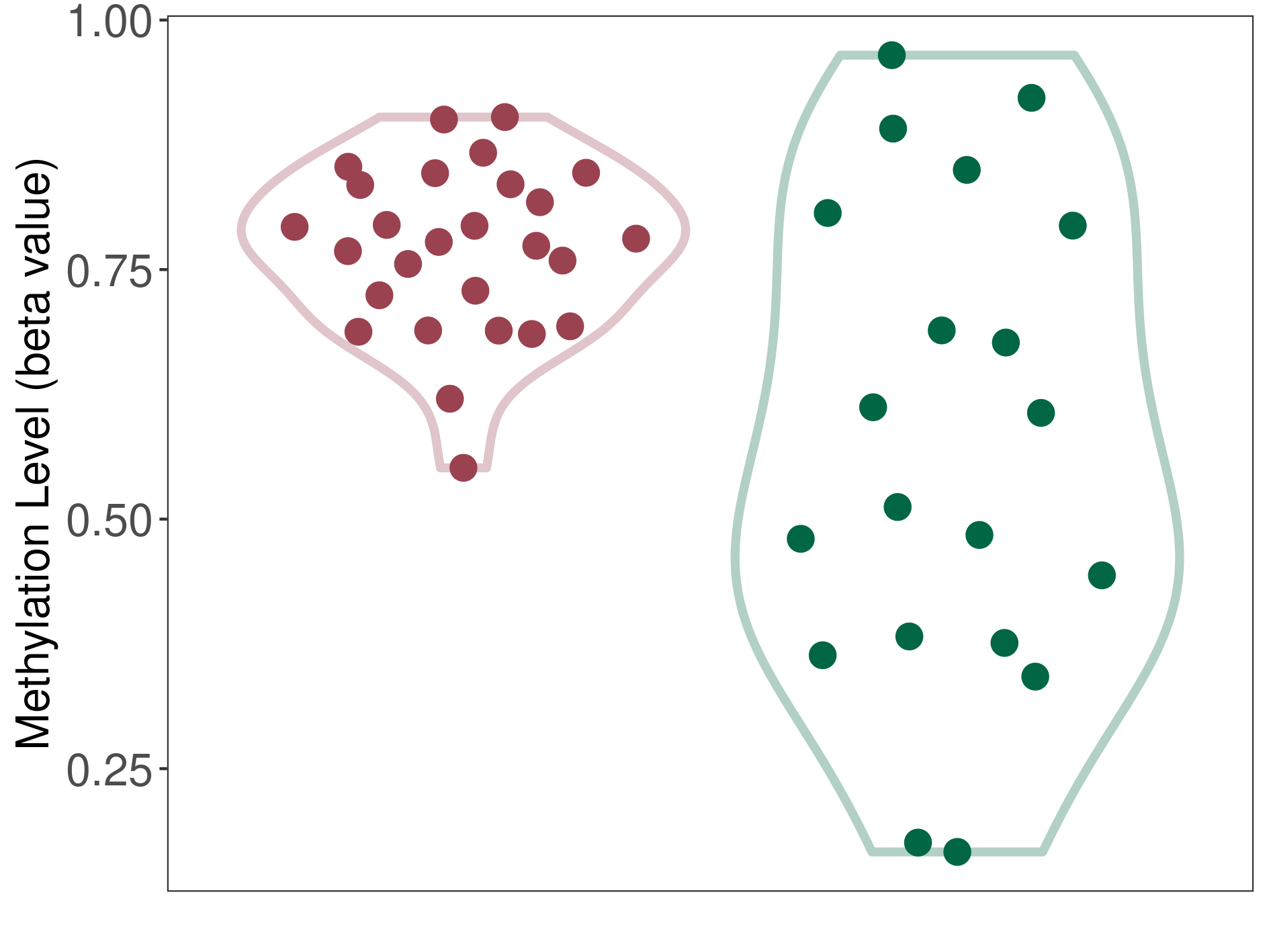
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
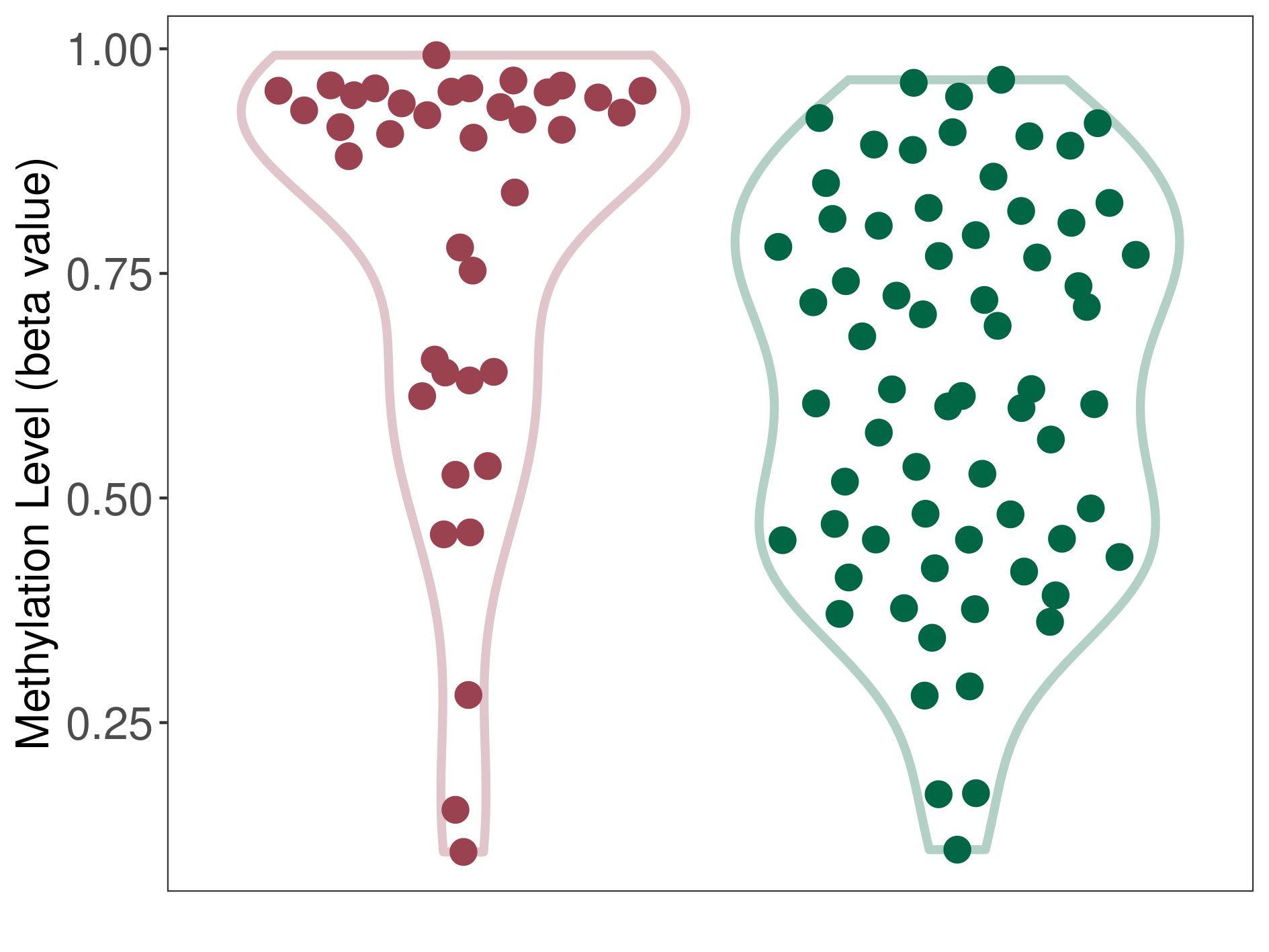
Lymphoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Moderate hypermethylation of CACNG2 in lymphoma than that in healthy individual | ||||
Studied Phenotype |
Lymphoma [ICD-11:2B30] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 0.001330689; Fold-change: 0.290423253; Z-score: 1.341719306 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
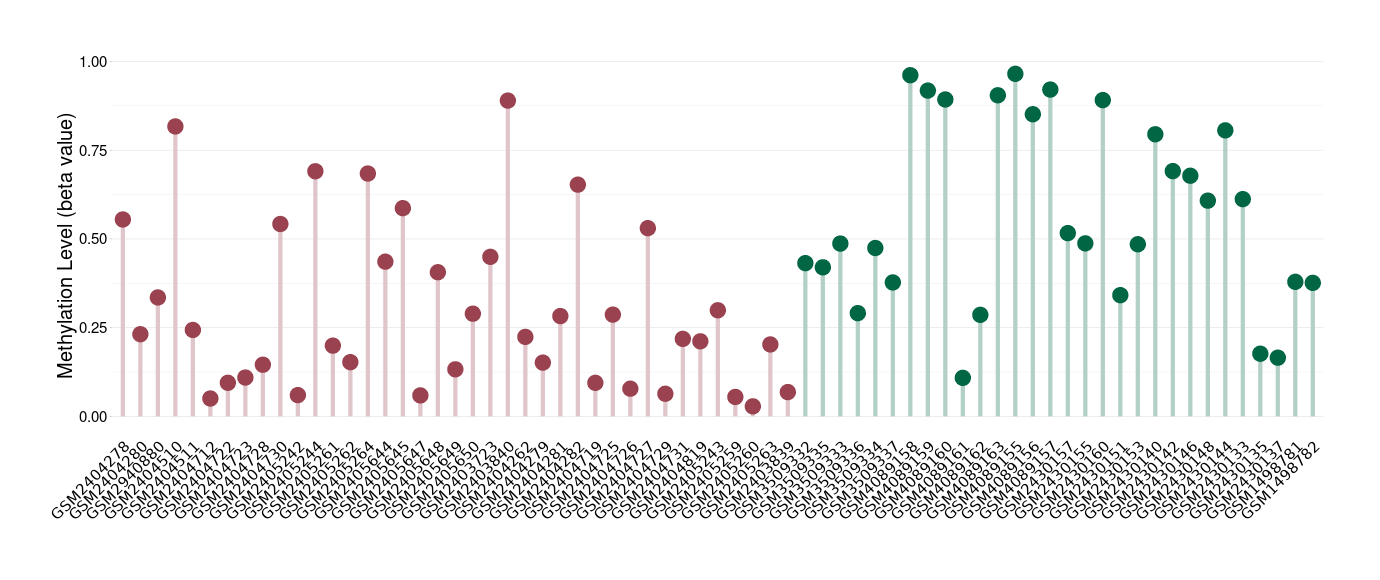
Esthesioneuroblastoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Moderate hypomethylation of CACNG2 in esthesioneuroblastoma than that in healthy individual | ||||
Studied Phenotype |
Esthesioneuroblastoma [ICD-11:2D50.1] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 2.55E-05; Fold-change: -0.2779795; Z-score: -1.054736877 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||

|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
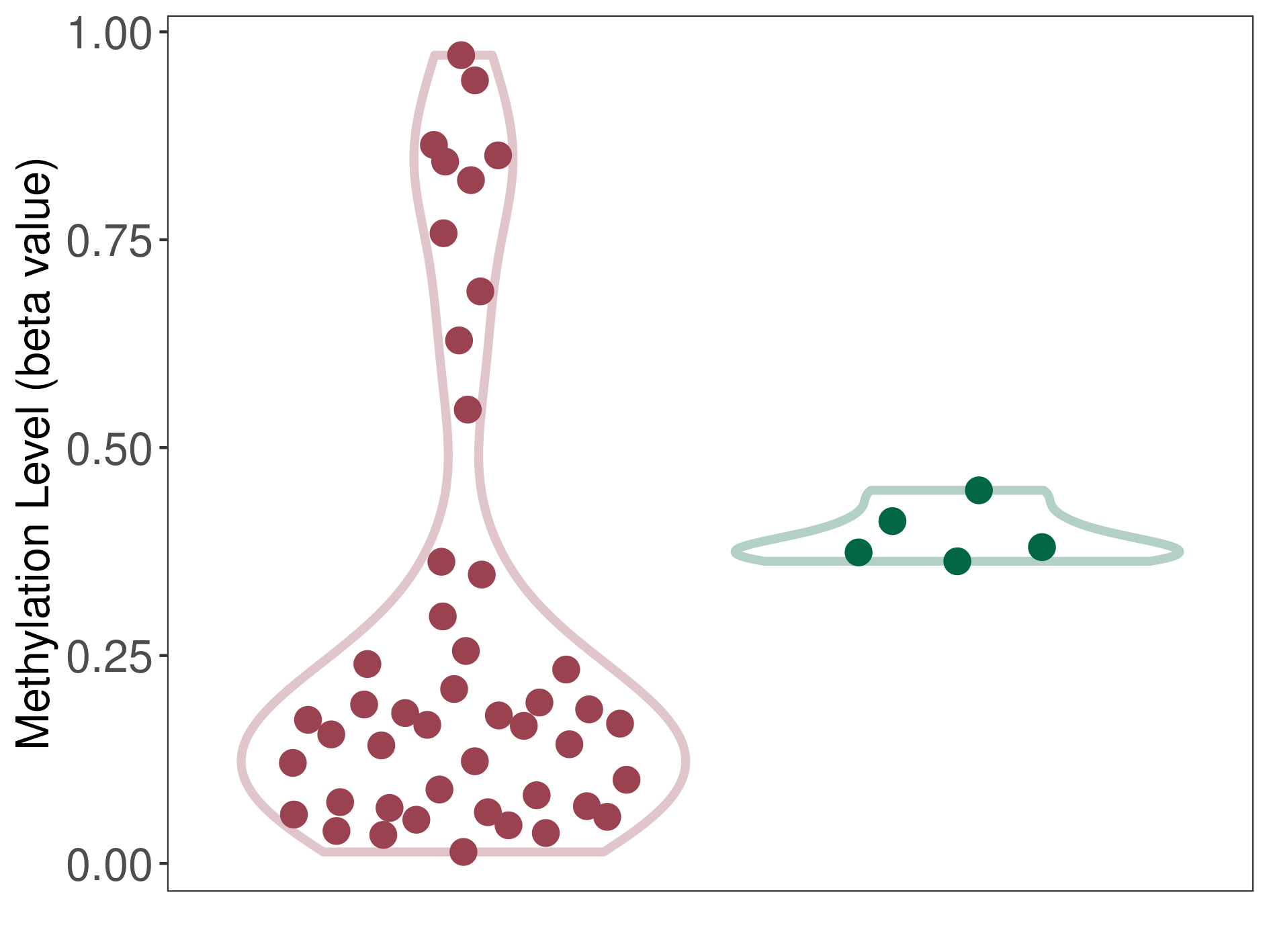
Oligodendroglial tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Moderate hypomethylation of CACNG2 in oligodendroglial tumour than that in healthy individual | ||||
Studied Phenotype |
Oligodendroglial tumour [ICD-11:2A00.Y1] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 0.016533483; Fold-change: -0.209984535; Z-score: -6.052861535 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Myxopapillary ependymoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
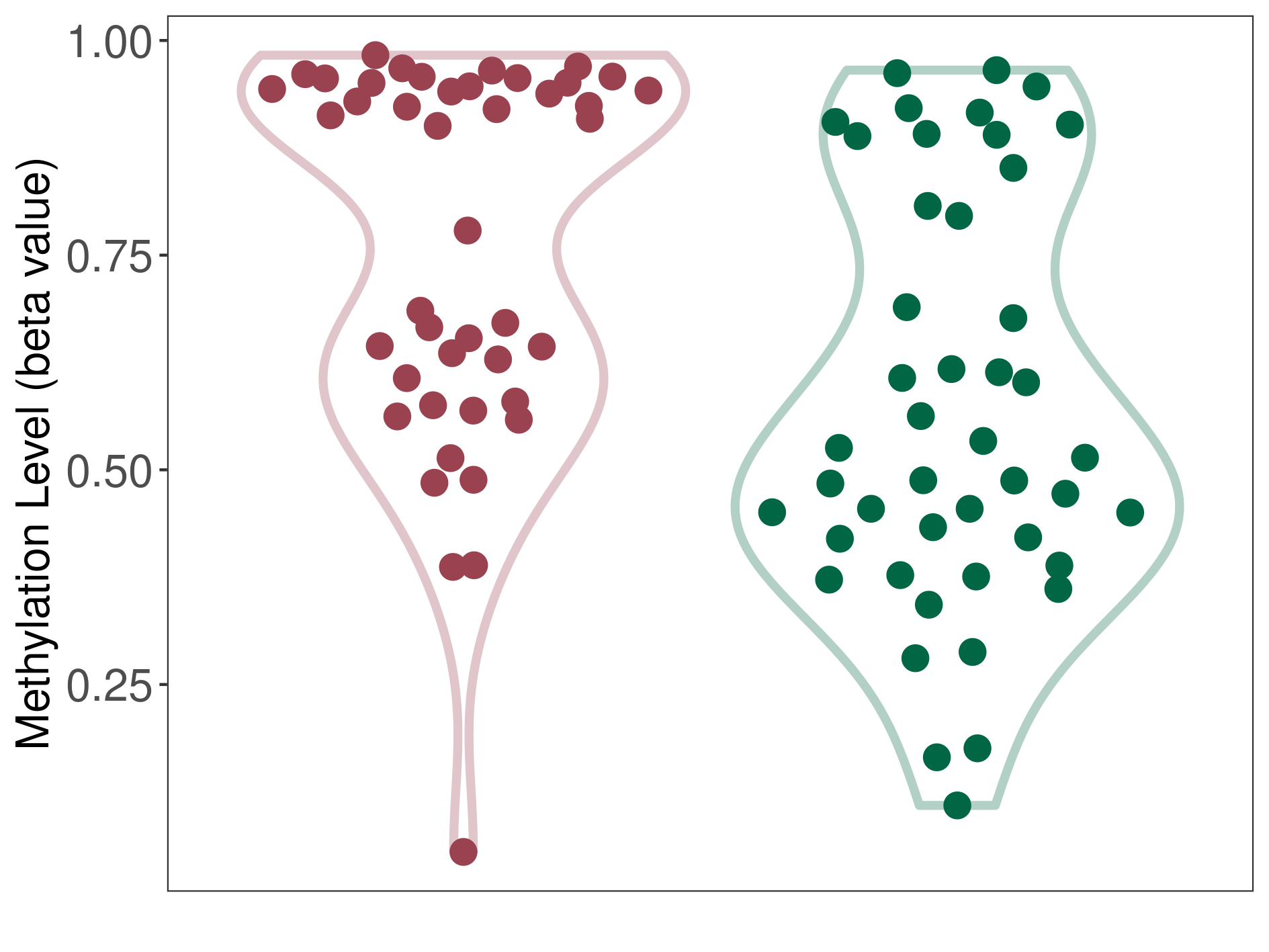
Epigenetic Phenomenon 1 |
Significant hypermethylation of CACNG2 in myxopapillary ependymoma than that in healthy individual | ||||
Studied Phenotype |
Myxopapillary ependymoma [ICD-11:2A00.5] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 0.000240329; Fold-change: 0.390543084; Z-score: 1.627029009 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Third ventricle chordoid glioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
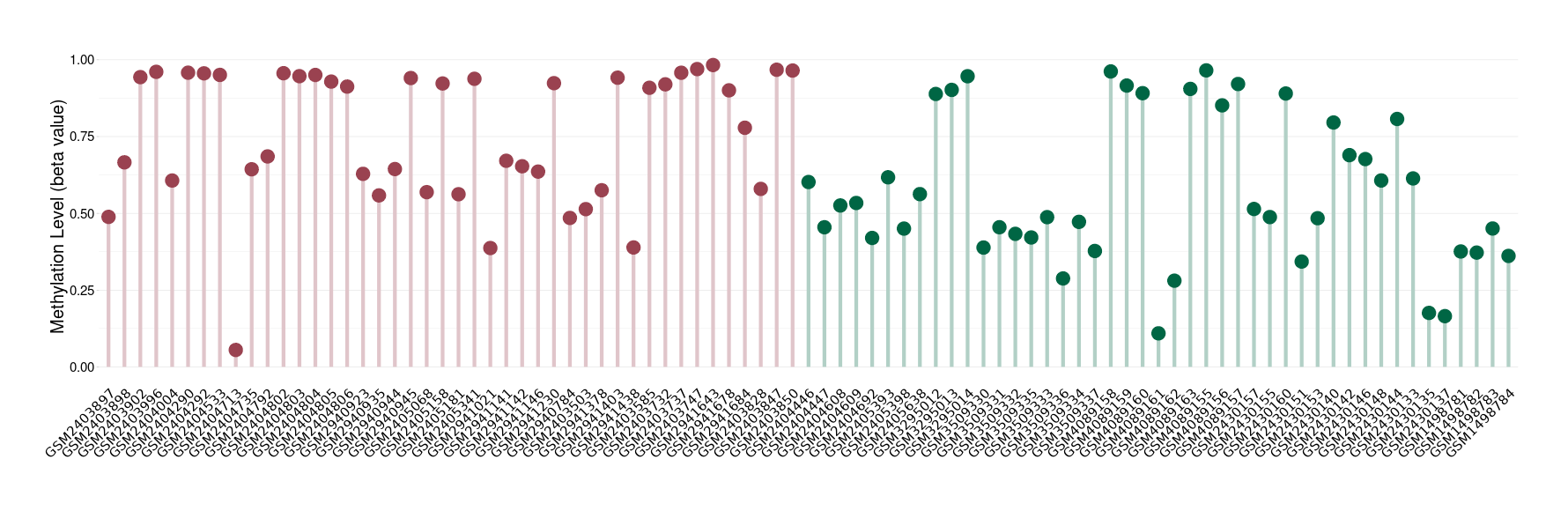
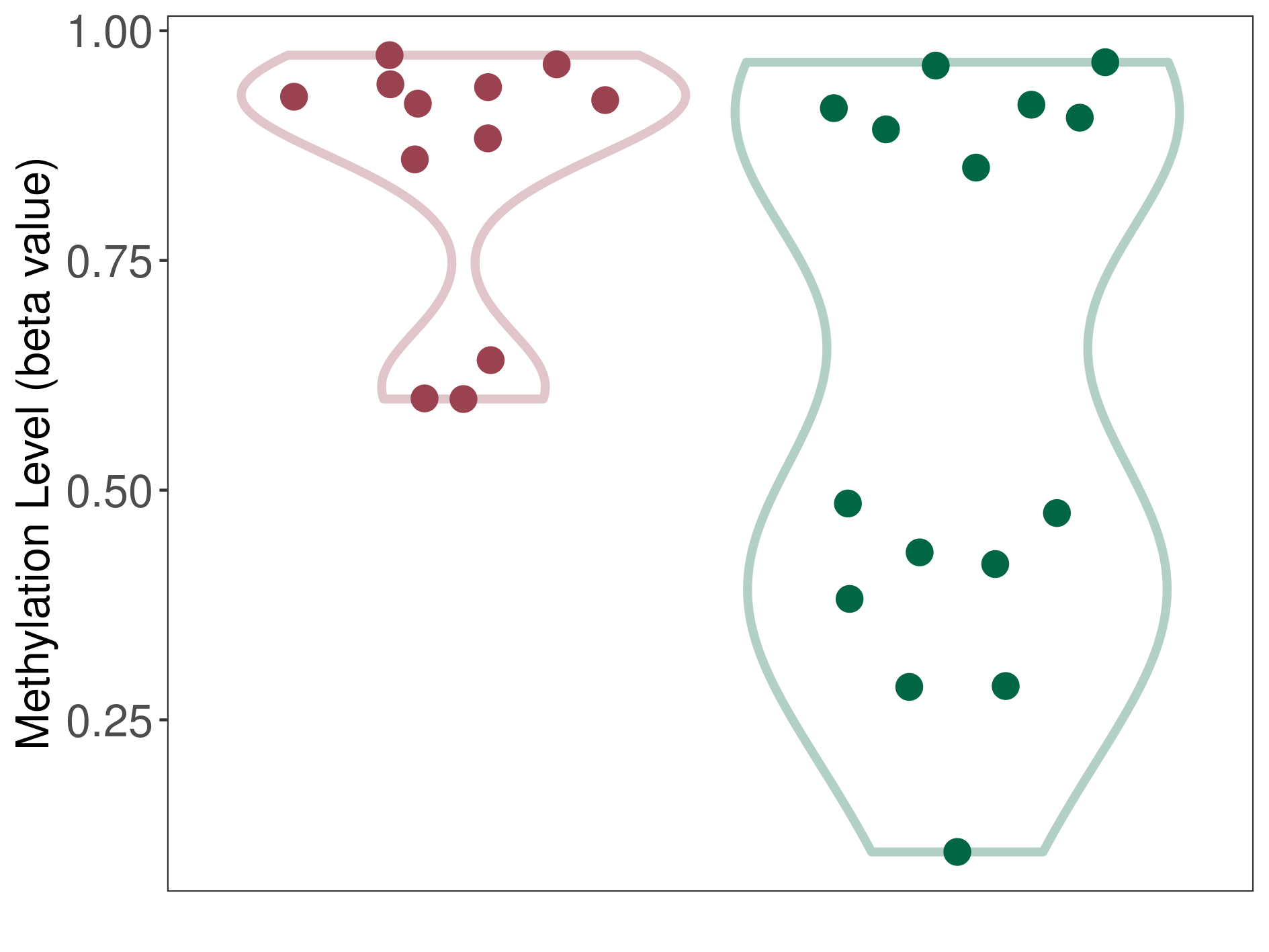
Epigenetic Phenomenon 1 |
Significant hypermethylation of CACNG2 in third ventricle chordoid glioma than that in healthy individual | ||||
Studied Phenotype |
Third ventricle chordoid glioma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 0.017365893; Fold-change: 0.437102162; Z-score: 1.445184583 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Brain neuroblastoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
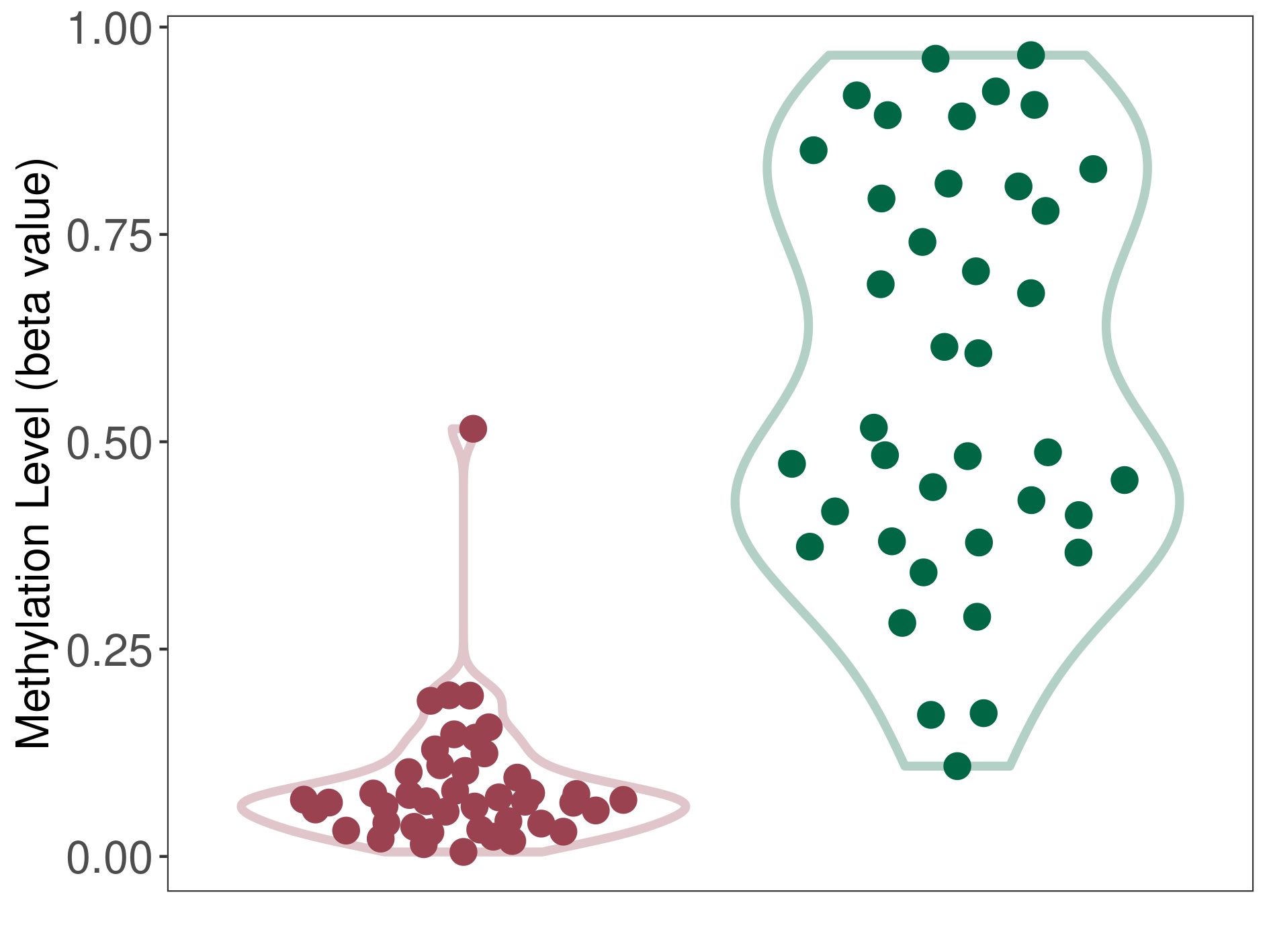
Epigenetic Phenomenon 1 |
Significant hypomethylation of CACNG2 in brain neuroblastoma than that in healthy individual | ||||
Studied Phenotype |
Brain neuroblastoma [ICD-11:2A00.11] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 1.00E-15; Fold-change: -0.450509159; Z-score: -1.820253049 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
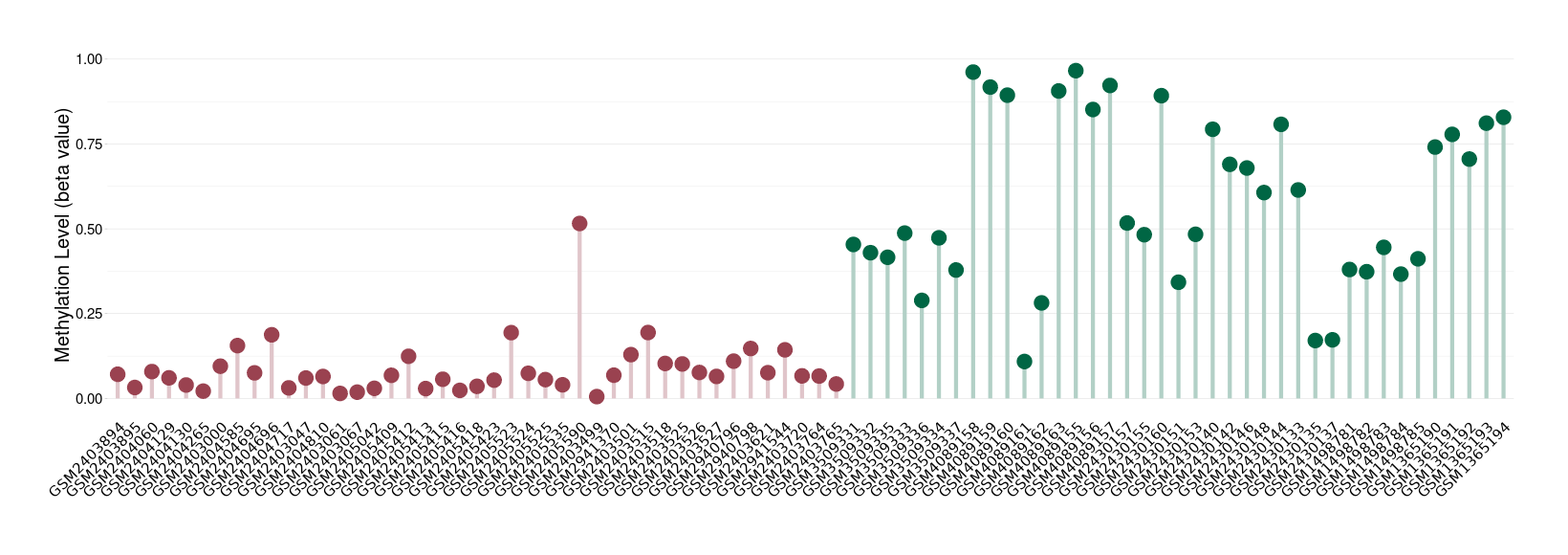 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Central neurocytoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypomethylation of CACNG2 in central neurocytoma than that in healthy individual | ||||
Studied Phenotype |
Central neurocytoma [ICD-11:2A00.3] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 4.04E-06; Fold-change: -0.505035521; Z-score: -1.821621963 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
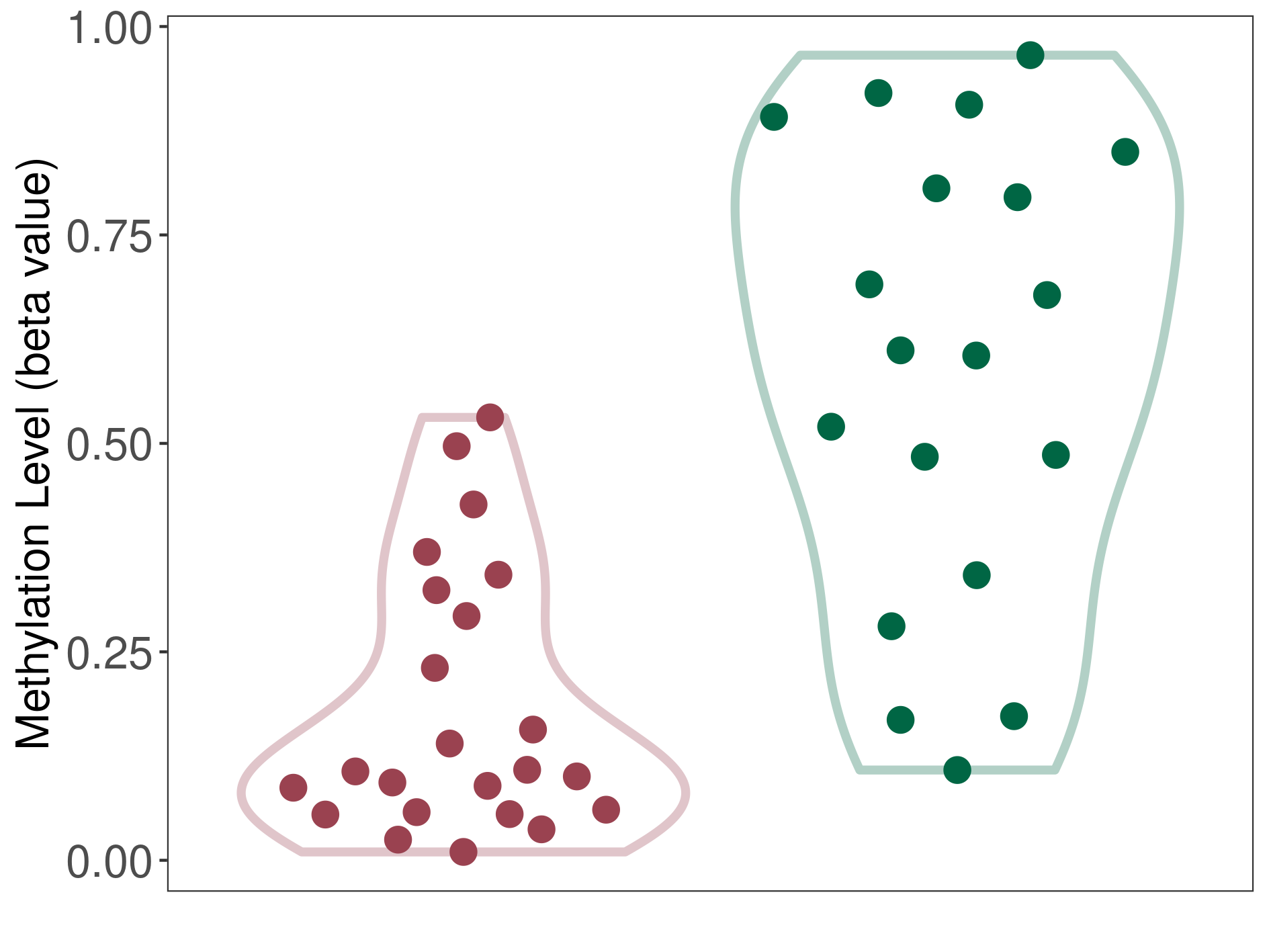
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
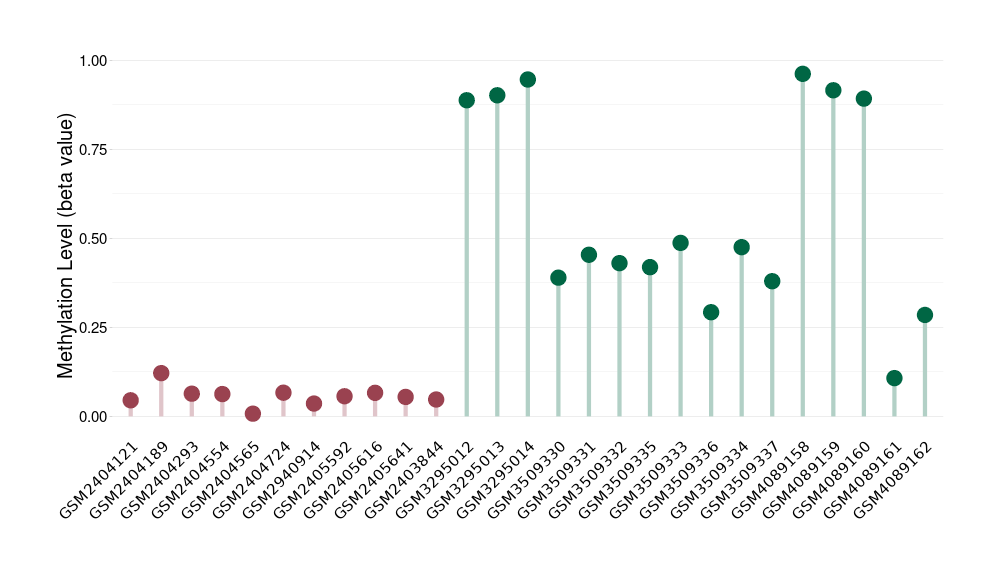
Cerebellar liponeurocytoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypomethylation of CACNG2 in cerebellar liponeurocytoma than that in healthy individual | ||||
Studied Phenotype |
Cerebellar liponeurocytoma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 2.76E-06; Fold-change: -0.407967686; Z-score: -1.417846546 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
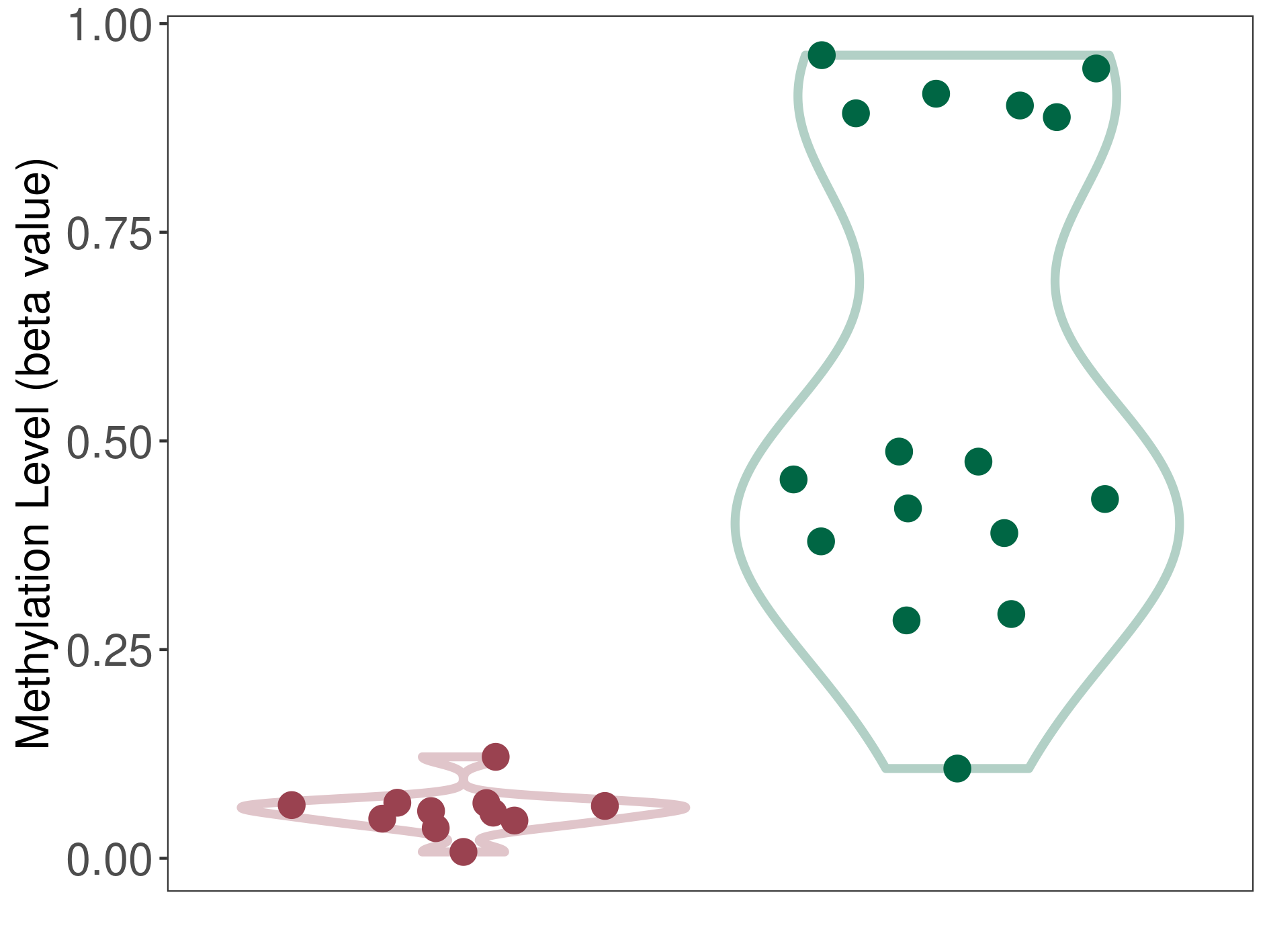
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
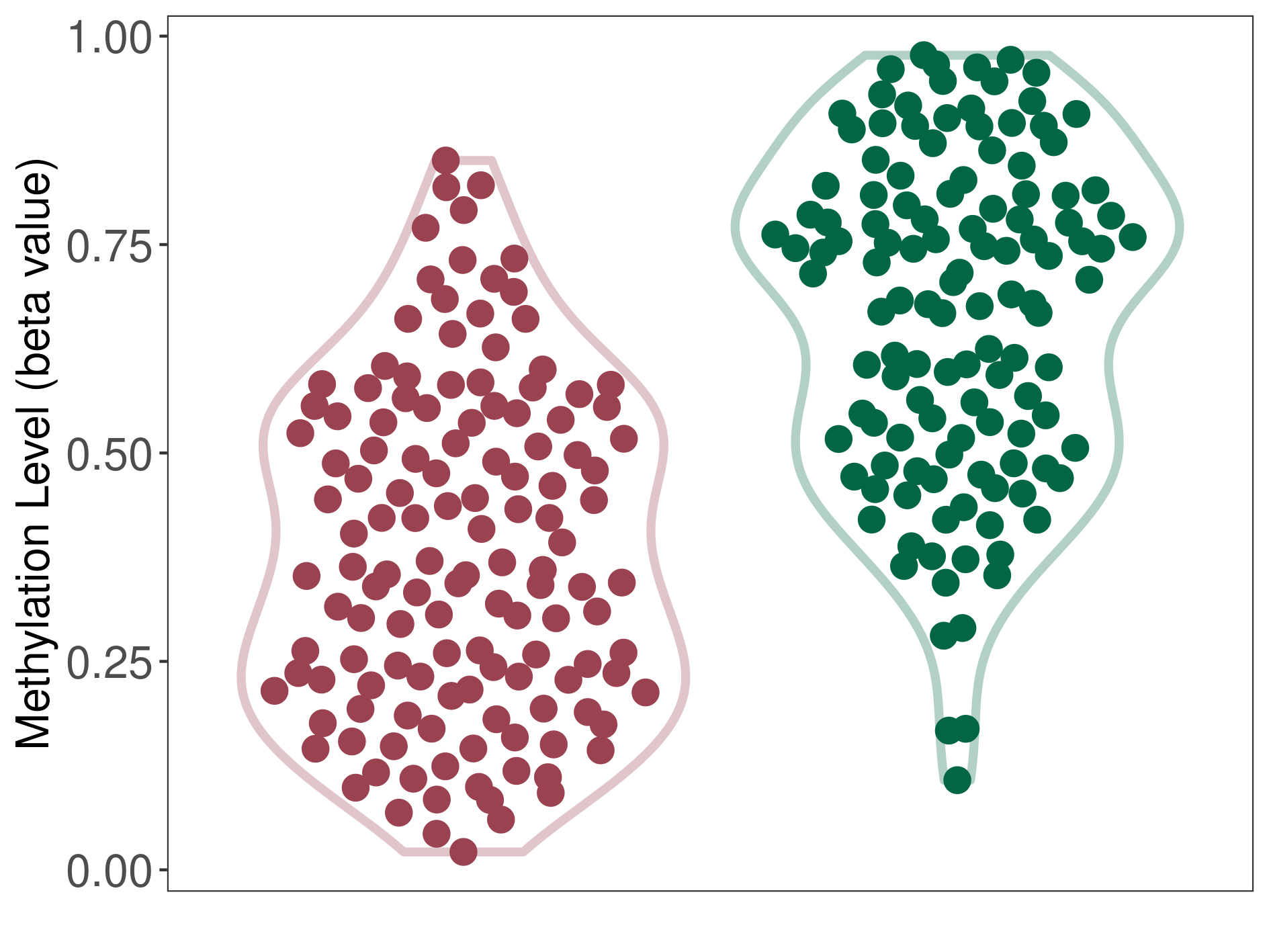
Diffuse midline glioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypomethylation of CACNG2 in diffuse midline glioma than that in healthy individual | ||||
Studied Phenotype |
Diffuse midline glioma [ICD-11:2A00.0Z] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 1.85E-24; Fold-change: -0.349307513; Z-score: -1.763618271 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
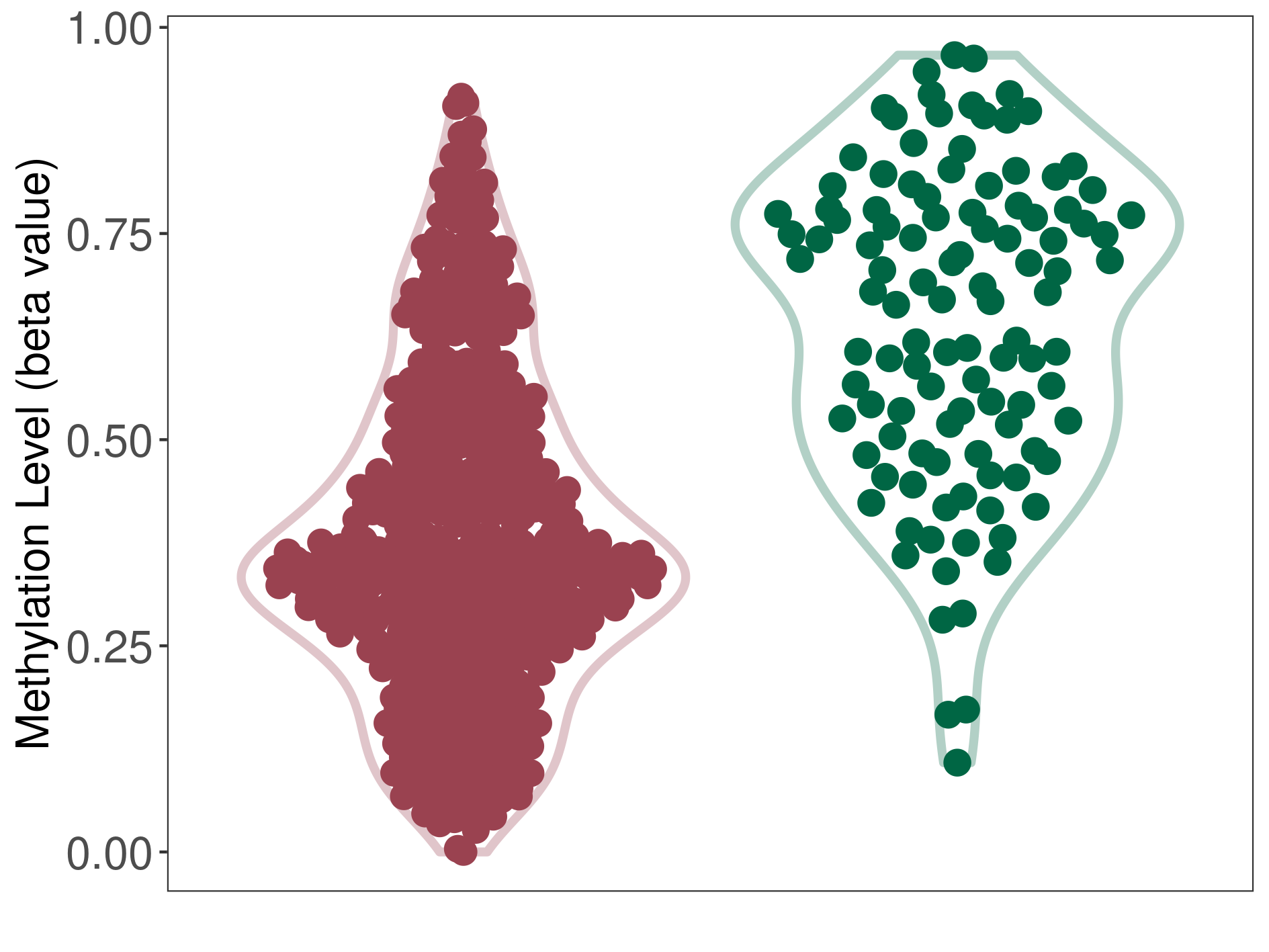
Low grade glioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypomethylation of CACNG2 in low grade glioma than that in healthy individual | ||||
Studied Phenotype |
Low grade glioma [ICD-11:2A00.0Z] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 5.18E-27; Fold-change: -0.329267941; Z-score: -1.741185023 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
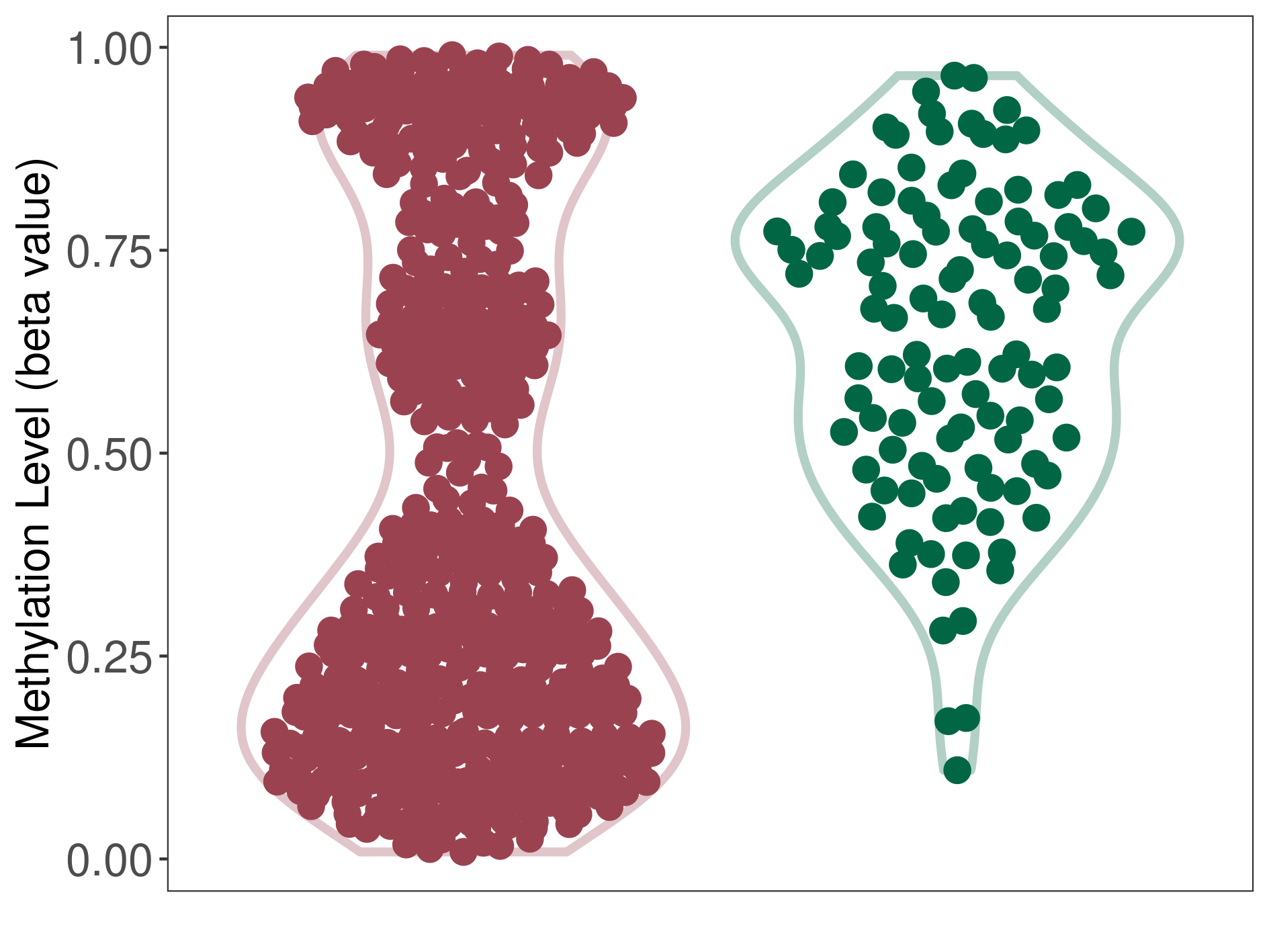
Malignant astrocytoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypomethylation of CACNG2 in malignant astrocytoma than that in healthy individual | ||||
Studied Phenotype |
Malignant astrocytoma [ICD-11:2A00.12] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 8.98E-15; Fold-change: -0.322497683; Z-score: -1.707798997 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Medulloblastoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypomethylation of CACNG2 in medulloblastoma than that in healthy individual | ||||
Studied Phenotype |
Medulloblastoma [ICD-11:2A00.10] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 4.84E-51; Fold-change: -0.600352582; Z-score: -2.834401359 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
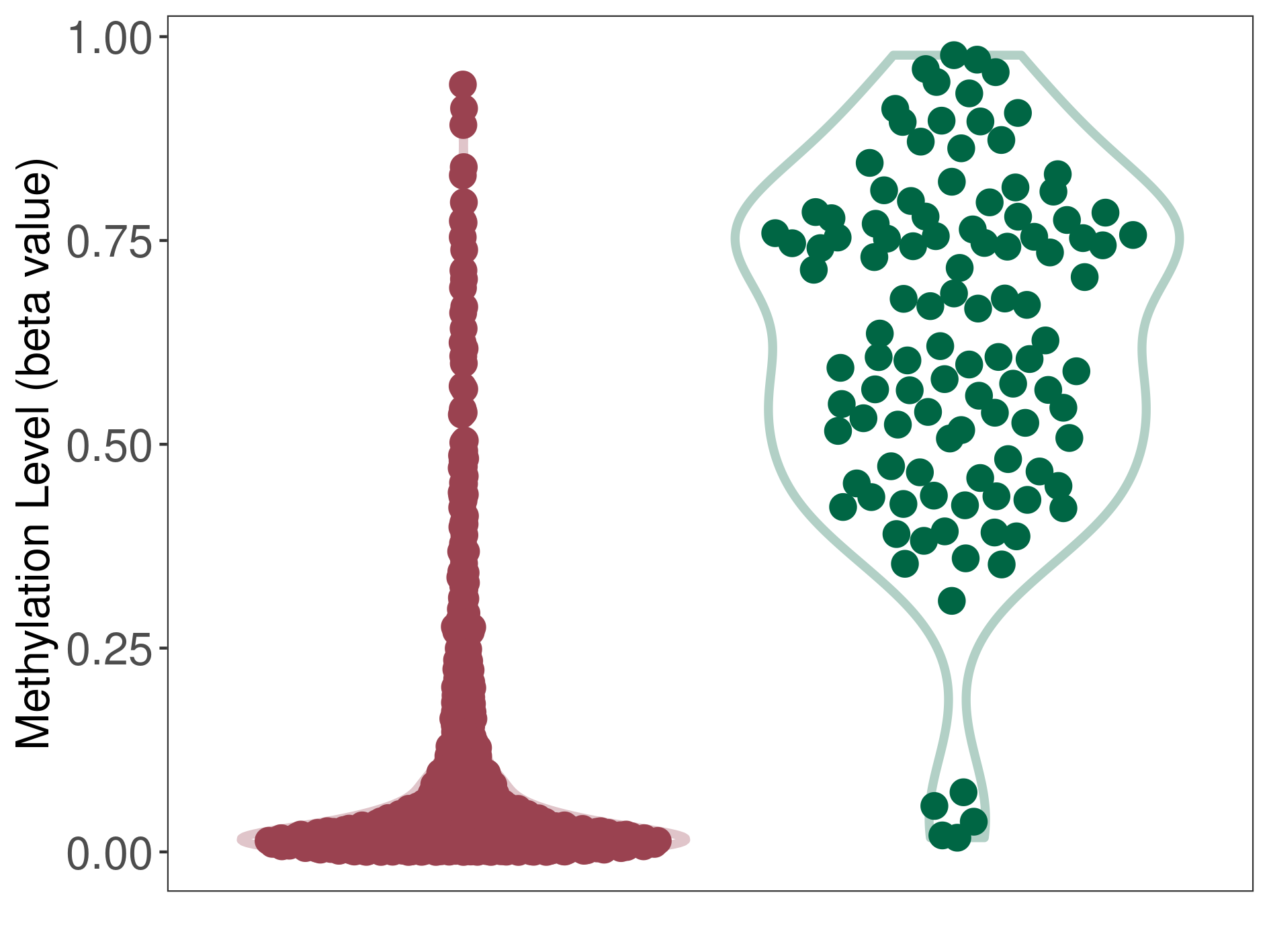
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Mixed neuronal-glial tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypomethylation of CACNG2 in mixed neuronal-glial tumour than that in healthy individual | ||||
Studied Phenotype |
Mixed neuronal-glial tumour [ICD-11:2A00.21] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 4.86E-15; Fold-change: -0.332964324; Z-score: -1.441868379 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
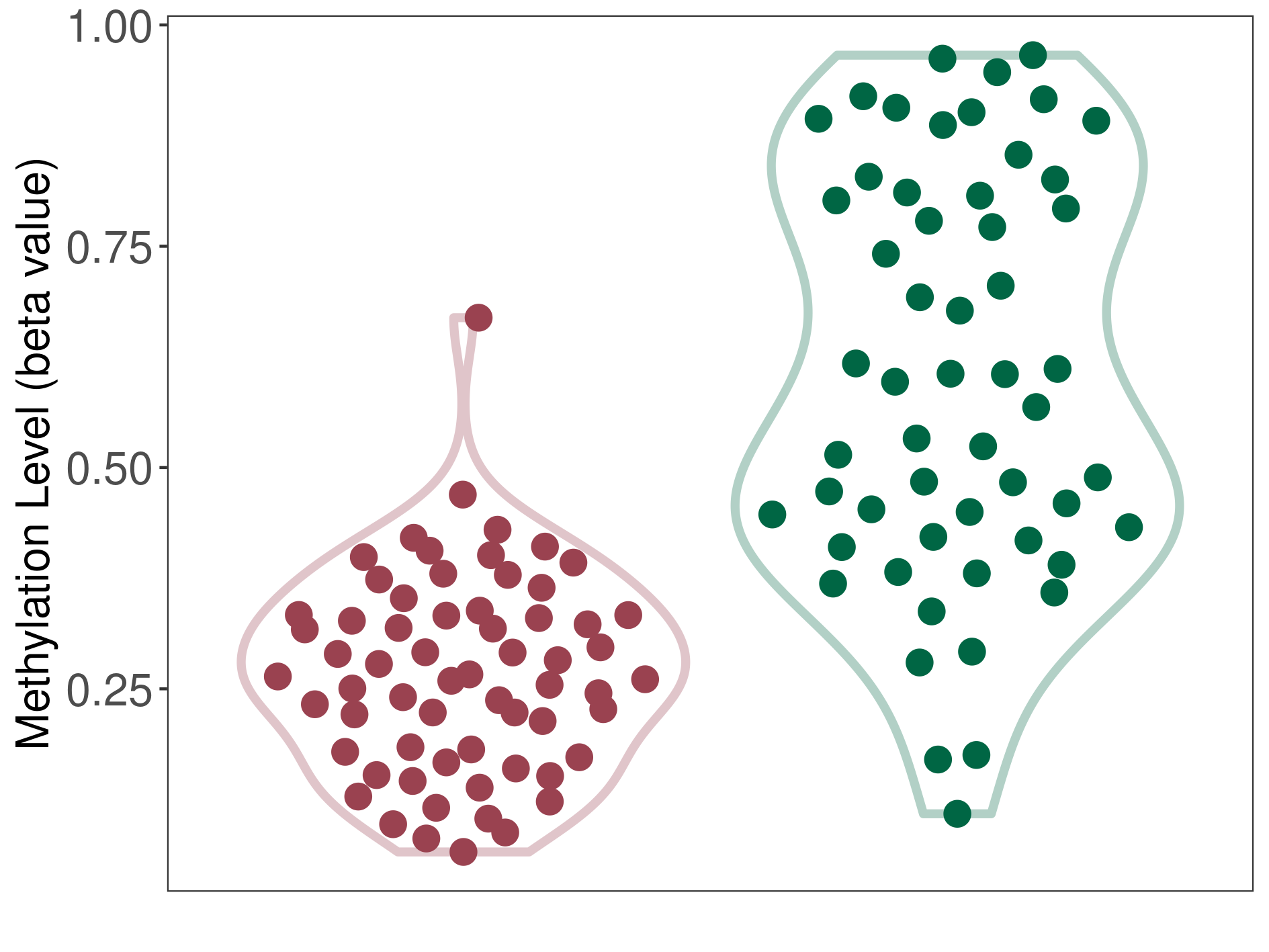
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
|
Oligodendroglioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon 1 |
Significant hypomethylation of CACNG2 in oligodendroglioma than that in healthy individual | ||||
Studied Phenotype |
Oligodendroglioma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value: 4.07E-29; Fold-change: -0.511137634; Z-score: -2.359240886 | ||||
|
DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
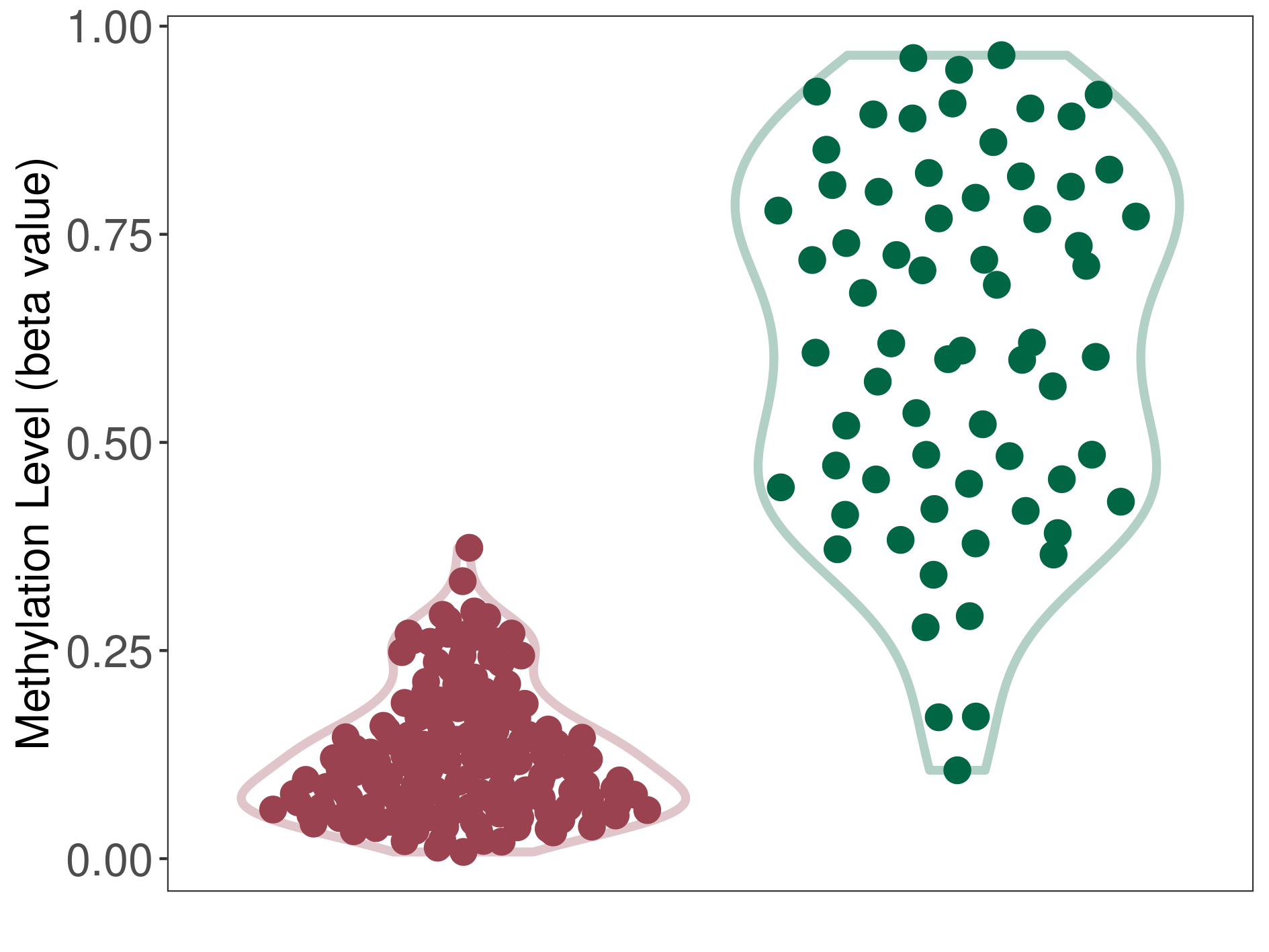
|
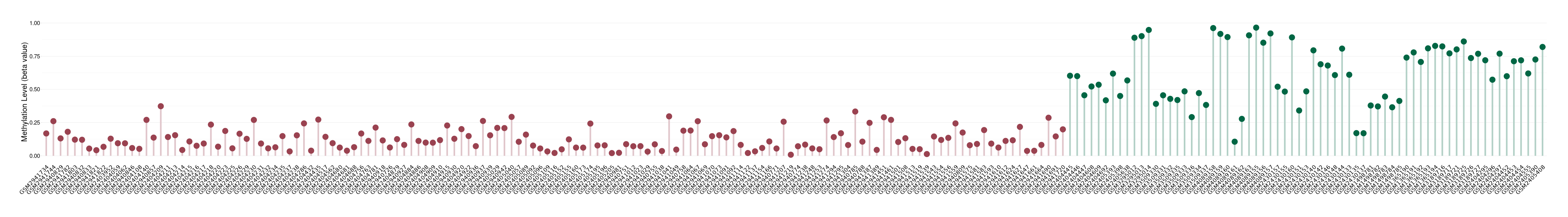 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
|
||||
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.
