Modelled Stucture
Method: Homology modeling
Teplate PDB: 6JMQ_A
Identity: 49.281%
Minimized Score: -1068.465
Detail: Structure Info
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0473 Transporter Info | ||||
| Gene Name | SLC7A6 | ||||
| Protein Name | Y+L amino acid transporter 2 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| 3D Structure |
Modelled Stucture Method: Homology modeling Teplate PDB: 6JMQ_A Identity: 49.281% Minimized Score: -1068.465 Detail: Structure Info |
||||
| Inter-species Structural Differences (ISD) | |||||
| Mus musculus (Mouse) | |||||
| Gene Name | Slc7a6 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method: Homology modeling Teplate PDB: 7CCS_B Sequence Length: 484 Identity: 55.992% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -1059.084 kcal/mol | |||
| Ramachandra Favored |
Medium |
||||
| QMEANBrane Quality |
Medium |
||||
| Ramachandran Plot |

Ramz Z Score: -1.87 ± 0.34 Residues in Favored Region: 458 Ramachandran favored: 95.02% Number of Outliers: 5 Ramachandran outliers: 1.04% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
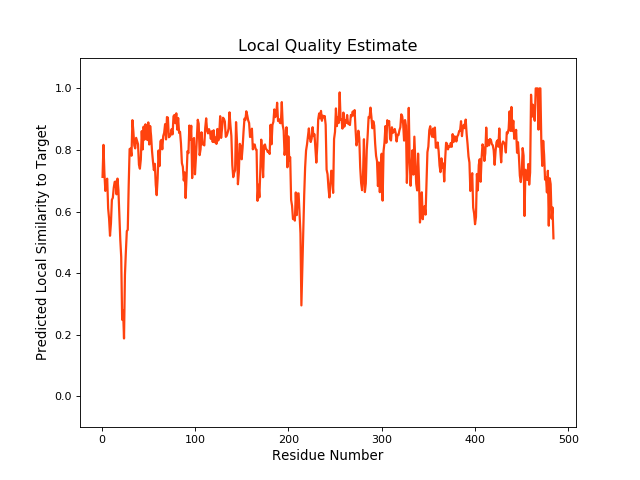
QMEANBrane Score: 0.79 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Inter-species Structural Differences (ISD) | |||||
| Danio rerio (Zebrafish) | |||||
| Gene Name | slc7a6 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method: Homology modeling Teplate PDB: 7CCS_B Sequence Length: 454 Identity: 58.37% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -1103.323 kcal/mol | |||
| Ramachandra Favored |
Poor |
||||
| QMEANBrane Quality |
Medium |
||||
| Ramachandran Plot |

Ramz Z Score: -1.12 ± 0.37 Residues in Favored Region: 429 Ramachandran favored: 94.91% Number of Outliers: 6 Ramachandran outliers: 1.33% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |

QMEANBrane Score: 0.79 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Inter-species Structural Differences (ISD) | |||||
| Xenopus laevis (African clawed frog) | |||||
| Gene Name | slc7a6 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method: Homology modeling Teplate PDB: 7CCS_B Sequence Length: 466 Identity: 54.936% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -1074.22 kcal/mol | |||
| Ramachandra Favored |
Medium |
||||
| QMEANBrane Quality |
Medium |
||||
| Ramachandran Plot |

Ramz Z Score: -1.83 ± 0.33 Residues in Favored Region: 443 Ramachandran favored: 95.47% Number of Outliers: 4 Ramachandran outliers: 0.86% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
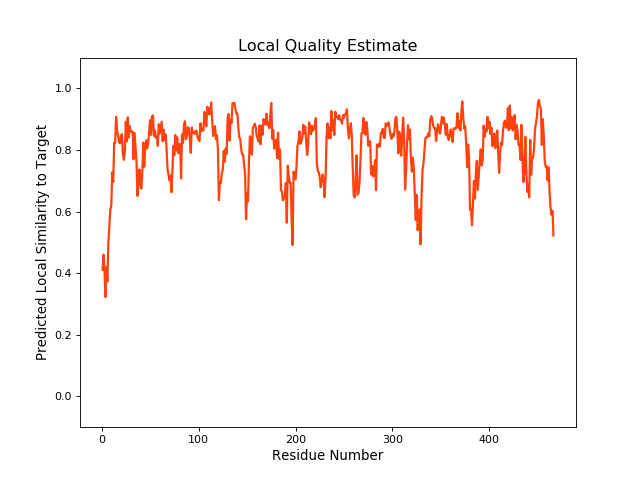
QMEANBrane Score: 0.79 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.