Modelled Stucture
Method: Homology modeling
Teplate PDB: 3US9_A
Identity: 87.888%
Minimized Score: -743.553
Detail: Structure Info
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0477 Transporter Info | ||||
| Gene Name | SLC8A1 | ||||
| Protein Name | Sodium/calcium exchanger 1 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| 3D Structure |
Modelled Stucture Method: Homology modeling Teplate PDB: 3US9_A Identity: 87.888% Minimized Score: -743.553 Detail: Structure Info |
||||
| Inter-species Structural Differences (ISD) | |||||
| Mus musculus (Mouse) | |||||
| Gene Name | Slc8a1 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method: Homology modeling Teplate PDB: 3US9_A Sequence Length: 322 Identity: 83.23% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -717.951 kcal/mol | |||
| Ramachandra Favored |
Medium |
||||
| QMEANBrane Quality |
Medium |
||||
| Ramachandran Plot |

Ramz Z Score: 0.26 ± 0.47 Residues in Favored Region: 306 Ramachandran favored: 95.62% Number of Outliers: 3 Ramachandran outliers: 0.94% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |

QMEANBrane Score: 0.75 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Inter-species Structural Differences (ISD) | |||||
| Rattus norvegicus (Rat) | |||||
| Gene Name | Slc8a1 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method: Homology modeling Teplate PDB: 3US9_A Sequence Length: 323 Identity: 84.83% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -764.469 kcal/mol | |||
| Ramachandra Favored |
Poor |
||||
| QMEANBrane Quality |
Medium |
||||
| Ramachandran Plot |

Ramz Z Score: -0.77 ± 0.45 Residues in Favored Region: 304 Ramachandran favored: 94.70% Number of Outliers: 7 Ramachandran outliers: 2.18% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |

QMEANBrane Score: 0.75 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Inter-species Structural Differences (ISD) | |||||
| Canis lupus familiaris (Dog) | |||||
| Gene Name | SLC8A1 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method: Homology modeling Teplate PDB: 3US9_A Sequence Length: 322 Identity: 88.82% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -720.531 kcal/mol | |||
| Ramachandra Favored |
Poor |
||||
| QMEANBrane Quality |
Medium |
||||
| Ramachandran Plot |
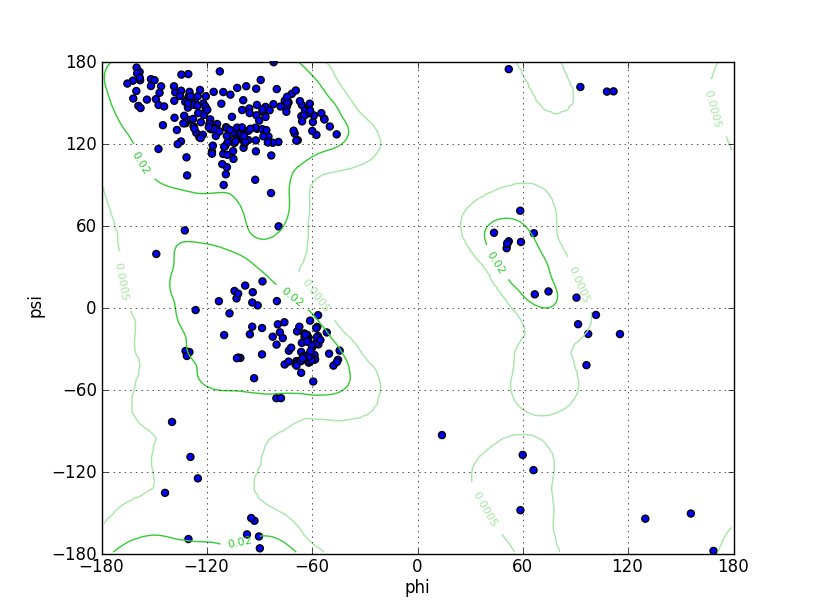
Ramz Z Score: 0.13 ± 0.45 Residues in Favored Region: 302 Ramachandran favored: 94.38% Number of Outliers: 2 Ramachandran outliers: 0.62% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |

QMEANBrane Score: 0.75 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Experimental Structures of This Model Oganism | |||||
| PDB ID | Scanning Method | Resolution | Expression System | Details | Ref |
| 2FWS | NMR | N.A. | Escherichia coli | [ 1] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A | ||||
| Sequence Length | 403-541 | Mutation | No | ||
| 2FWU | NMR | N.A. | Escherichia coli | [ 1] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A | ||||
| Sequence Length | 533-724 | Mutation | No | ||
| 2DPK | X-ray | 2.5 Å | N.A. | [ 2] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A | ||||
| Sequence Length | 402-541 | Mutation | No | ||
| 2QVK | X-ray | 1.45 Å | N.A. | [ 3] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A | ||||
| Sequence Length | 533-721 | Mutation | No | ||
| 2QVM | X-ray | 1.7 Å | N.A. | [ 3] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A | ||||
| Sequence Length | 533-721 | Mutation | No | ||
| 3GIN | X-ray | 2.4 Å | Escherichia coli | [ 4] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A/B | ||||
| Sequence Length | 402-541 | Mutation | Yes | ||
| 3US9 | X-ray | 2.68 Å | Escherichia coli | [ 5] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A | ||||
| Sequence Length | 403-724 | Mutation | Yes | ||
| 6BV7 | NMR | N.A. | Escherichia coli | [ 6] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A | ||||
| Sequence Length | 306-359 | Mutation | No | ||
| References | |||||
| 1 | Ca2+ regulation in the Na+/Ca2+ exchanger involves two markedly different Ca2+ sensors. Mol Cell. 2006 Apr 7;22(1):15-25. | ||||
| 2 | The crystal structure of the primary Ca2+ sensor of the Na+/Ca2+ exchanger reveals a novel Ca2+ binding motif. J Biol Chem. 2006 Aug 4;281(31):21577-21581. | ||||
| 3 | The second Ca2+-binding domain of the Na+ Ca2+ exchanger is essential for regulation: crystal structures and mutational analysis. Proc Natl Acad Sci U S A. 2007 Nov 20;104(47):18467-72. | ||||
| 4 | Structure and functional analysis of a Ca2+ sensor mutant of the Na+/Ca2+ exchanger. J Biol Chem. 2009 May 29;284(22):14688-92. | ||||
| 5 | A common Ca2+-driven interdomain module governs eukaryotic NCX regulation. PLoS One. 2012;7(6):e39985. | ||||
| 6 | The Intracellular Loop of the Na(+)/Ca(2+) Exchanger Contains an "Awareness Ribbon"-Shaped Two-Helix Bundle Domain. Biochemistry. 2018 Aug 28;57(34):5096-5104. | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.