Modelled Stucture
Method: Homology modeling
Teplate PDB: 2W6H_I
Identity: 94.118%
Minimized Score: -75.374
Detail: Structure Info
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0517 Transporter Info | ||||
| Gene Name | ATP5E | ||||
| Protein Name | ATP synthase subunit epsilon | ||||
| Gene ID | |||||
| UniProt ID | |||||
| 3D Structure |
Modelled Stucture Method: Homology modeling Teplate PDB: 2W6H_I Identity: 94.118% Minimized Score: -75.374 Detail: Structure Info |
||||
| Inter-species Structural Differences (ISD) | |||||
| Mus musculus (Mouse) | |||||
| Gene Name | Atp5f1e | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method: Homology modeling Teplate PDB: 6TT7_I Sequence Length: 51 Identity: 92.157% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -89.215 kcal/mol | |||
| Ramachandra Favored |
Medium |
||||
| QMEANBrane Quality |
Medium |
||||
| Ramachandran Plot |

Ramz Z Score: -2.45 ± 0.98 Residues in Favored Region: 48 Ramachandran favored: 97.96% Number of Outliers: 0 Ramachandran outliers: 0.00% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |

QMEANBrane Score: 0.73 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Inter-species Structural Differences (ISD) | |||||
| Rattus norvegicus (Rat) | |||||
| Gene Name | Atp5f1e | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method: Homology modeling Teplate PDB: 6TT7_I Sequence Length: 51 Identity: 90.196% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -77.843 kcal/mol | |||
| Ramachandra Favored |
Medium |
||||
| QMEANBrane Quality |
Medium |
||||
| Ramachandran Plot |
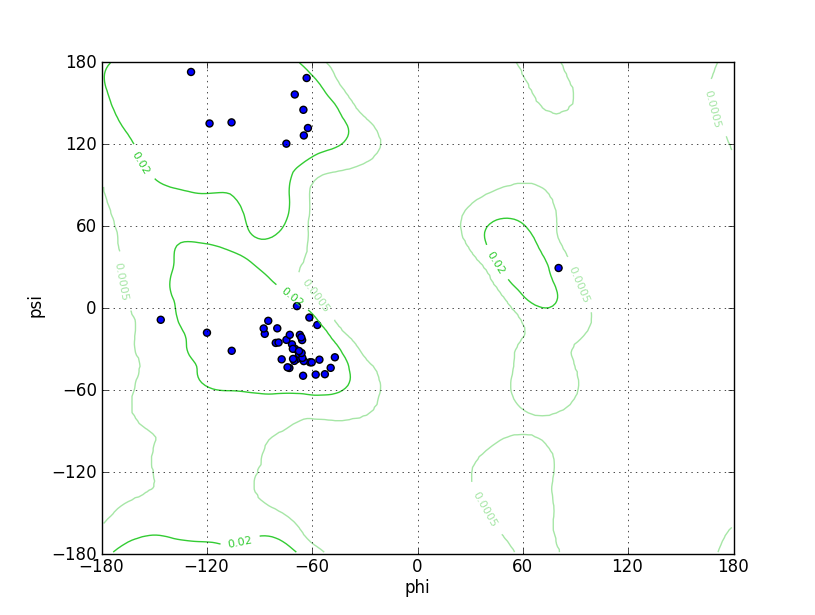
Ramz Z Score: -3.23 ± 0.98 Residues in Favored Region: 48 Ramachandran favored: 97.96% Number of Outliers: 0 Ramachandran outliers: 0.00% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |

QMEANBrane Score: 0.73 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Inter-species Structural Differences (ISD) | |||||
| Bos taurus (Bovine) | |||||
| Gene Name | ATP5F1E | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure | |||||
| Click to Save PDB File in PDB Format | |||||
| Experimental Structures of This Model Oganism | |||||
| PDB ID | Scanning Method | Resolution | Expression System | Details | Ref |
| 1H8E | X-ray | 2 Å | N.A. | [ 1] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | I | ||||
| Sequence Length | 2-51 | Mutation | Yes | ||
| 2CK3 | X-ray | 1.95 Å | N.A. | [ 2] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | I | ||||
| Sequence Length | 2-51 | Mutation | No | ||
| 2V7Q | X-ray | 2.1 Å | N.A. | [ 3] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | I | ||||
| Sequence Length | 2-51 | Mutation | No | ||
| 2W6H | X-ray | 5 Å | N.A. | [ 4] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | I | ||||
| Sequence Length | 1-51 | Mutation | No | ||
| 2W6I | X-ray | 4 Å | N.A. | [ 4] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | I | ||||
| Sequence Length | 1-51 | Mutation | No | ||
| 2W6J | X-ray | 3.84 Å | N.A. | [ 4] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | I | ||||
| Sequence Length | 1-51 | Mutation | No | ||
| 2WSS | X-ray | 3.2 Å | Escherichia coli | [ 5] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | I/R | ||||
| Sequence Length | 2-51 | Mutation | No | ||
| 2XND | X-ray | 3.5 Å | N.A. | [ 6] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | I | ||||
| Sequence Length | 2-48 | Mutation | No | ||
| References | |||||
| 1 | Structure of bovine mitochondrial F(1)-ATPase with nucleotide bound to all three catalytic sites: implications for the mechanism of rotary catalysis. Cell. 2001 Aug 10;106(3):331-41. | ||||
| 2 | How azide inhibits ATP hydrolysis by the F-ATPases. Proc Natl Acad Sci U S A. 2006 Jun 6;103(23):8646-9. | ||||
| 3 | How the regulatory protein, IF(1), inhibits F(1)-ATPase from bovine mitochondria. Proc Natl Acad Sci U S A. 2007 Oct 2;104(40):15671-6. | ||||
| 4 | Improving diffraction by humidity control: a novel device compatible with X-ray beamlines. Acta Crystallogr D Biol Crystallogr. 2009 Dec;65(Pt 12):1237-46. | ||||
| 5 | The structure of the membrane extrinsic region of bovine ATP synthase. Proc Natl Acad Sci U S A. 2009 Dec 22;106(51):21597-601. | ||||
| 6 | Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc Natl Acad Sci U S A. 2010 Sep 28;107(39):16823-7. | ||||
| 7 | Structural evidence of a new catalytic intermediate in the pathway of ATP hydrolysis by F1-ATPase from bovine heart mitochondria. Proc Natl Acad Sci U S A. 2012 Jul 10;109(28):11139-43. | ||||
| 8 | How release of phosphate from mammalian F1-ATPase generates a rotary substep. Proc Natl Acad Sci U S A. 2015 May 12;112(19):6009-14. | ||||
| 9 | Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM. Elife. 2015 Oct 6;4:e10180. | ||||
| 10 | Structure of the dimeric ATP synthase from bovine mitochondria. Proc Natl Acad Sci U S A. 2020 Sep 22;117(38):23519-23526. | ||||
| 11 | The structure of the central stalk in bovine F(1)-ATPase at 2.4 A resolution. Nat Struct Biol. 2000 Nov;7(11):1055-61. | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.