Crystal Structure of This DT
Crystallisation Method: X-ray
Corresponding Chain: A/B
Sequence Length: 41-315
Detail: Struture Info
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0545 Transporter Info | ||||
| Gene Name | KCNK2 | ||||
| Protein Name | Potassium channel subfamily K member 2 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| 3D Structure |
Crystal Structure of This DT Crystallisation Method: X-ray Corresponding Chain: A/B Sequence Length: 41-315 Detail: Struture Info |
||||
| Inter-species Structural Differences (ISD) | |||||
| Mus musculus (Mouse) | |||||
| Gene Name | Kcnk2 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure | |||||
| Click to Save PDB File in PDB Format | |||||
| Experimental Structures of This Model Oganism | |||||
| PDB ID | Scanning Method | Resolution | Expression System | Details | Ref |
| 6V36 | X-ray | 3.4 Å | Komagataella pastoris | [ 1] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A/B | ||||
| Sequence Length | 35-338 | Mutation | No | ||
| 6V3C | X-ray | 3.51 Å | Komagataella pastoris | [ 1] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A/B | ||||
| Sequence Length | 35-338 | Mutation | No | ||
| 6V3I | X-ray | 3.4 Å | Komagataella pastoris | [ 1] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A/B | ||||
| Sequence Length | 35-338 | Mutation | No | ||
| 6CQ6 | X-ray | 3.1 Å | Komagataella pastoris | [ 2] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A/B | ||||
| Sequence Length | 35-337 | Mutation | Yes | ||
| 6CQ8 | X-ray | 3 Å | Komagataella pastoris | [ 2] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A/B | ||||
| Sequence Length | 35-337 | Mutation | Yes | ||
| 6CQ9 | X-ray | 2.8 Å | Komagataella pastoris | [ 2] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A/B | ||||
| Sequence Length | 35-339 | Mutation | Yes | ||
| Inter-species Structural Differences (ISD) | |||||
| Rattus norvegicus (Rat) | |||||
| Gene Name | Kcnk2 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method: Homology modeling Teplate PDB: 6CQ6_A Sequence Length: 305 Identity: 93.77% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -672.849 kcal/mol | |||
| Ramachandra Favored |
Excellent |
||||
| QMEANBrane Quality |
High |
||||
| Ramachandran Plot |
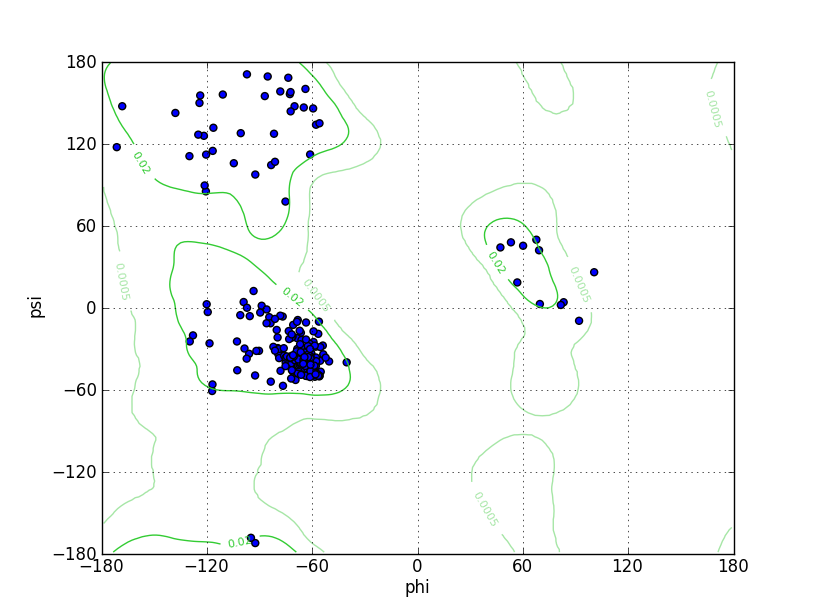
Ramz Z Score: 0.19 ± 0.43 Residues in Favored Region: 299 Ramachandran favored: 98.68% Number of Outliers: 0 Ramachandran outliers: 0.00% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
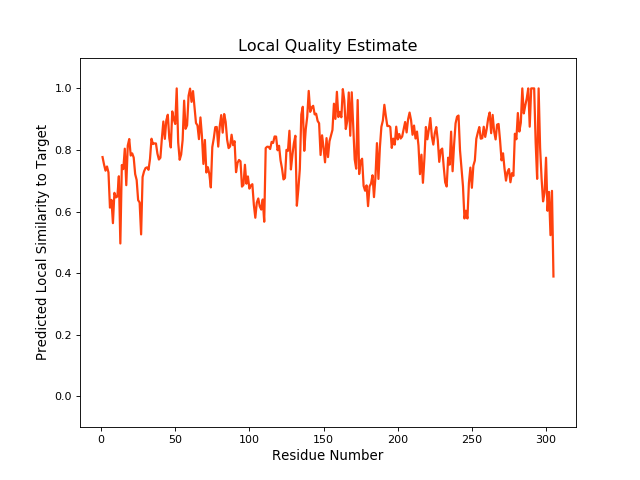
QMEANBrane Score: 0.8 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| References | |||||
| 1 | Polynuclear Ruthenium Amines Inhibit K(2P) Channels via a "Finger in the Dam" Mechanism. Cell Chem Biol. 2020 May 21;27(5):511-524.e4. | ||||
| 2 | K(2P)2.1 (TREK-1)-activator complexes reveal a cryptic selectivity filter binding site. Nature. 2017 Jul 20;547(7663):364-368. | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.