Modelled Stucture
Method: Loop building
Teplate PDB: 7A69_A
Identity: 99.74%
Minimized Score: -3251.1
Detail: Structure Info
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0003 Transporter Info | ||||
| Gene Name | ABCB1 | ||||
| Protein Name | P-glycoprotein 1 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| 3D Structure |
Modelled Stucture Method: Loop building Teplate PDB: 7A69_A Identity: 99.74% Minimized Score: -3251.1 Detail: Structure Info |
||||
| Inter-species Structural Differences (ISD) | |||||
| Mus musculus (Mouse) | |||||
| Gene Name | Abcb1a | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure | |||||
| Click to Save PDB File in PDB Format | |||||
| Experimental Structures of This Model Oganism | |||||
| PDB ID | Scanning Method | Resolution | Expression System | Details | Ref |
| 5KO2 | X-ray | 3.3 Å | Komagataella phaffii GS115 | [ 1] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A/B | ||||
| Sequence Length | 1-1276 | Mutation | Yes | ||
| 5KOY | X-ray | 3.85 Å | Komagataella phaffii GS116 | [ 1] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A/B | ||||
| Sequence Length | 1-1276 | Mutation | Yes | ||
| 5KPD | X-ray | 3.35 Å | Komagataella phaffii GS117 | [ 1] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A/B | ||||
| Sequence Length | 1-1276 | Mutation | Yes | ||
| 5KPI | X-ray | 4.01 Å | Komagataella phaffii GS118 | [ 1] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A/B | ||||
| Sequence Length | 1-1276 | Mutation | No | ||
| 3G5U | X-ray | 3.8 Å | N.A. | [ 2] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A/B | ||||
| Sequence Length | 1-1276 | Mutation | Yes | ||
| 3G60 | X-ray | 4.4 Å | N.A. | [ 2] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A/B | ||||
| Sequence Length | 1-1276 | Mutation | No | ||
| 3G61 | X-ray | 4.35 Å | N.A. | [ 2] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A/B | ||||
| Sequence Length | 1-1276 | Mutation | No | ||
| 4KSB | X-ray | 3.8 Å | Komagataella pastoris | [ 3] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A | ||||
| Sequence Length | 1-1276 | Mutation | No | ||
| Inter-species Structural Differences (ISD) | |||||
| Rattus norvegicus (Rat) | |||||
| Gene Name | Abcb1 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method: Homology modeling Teplate PDB: 6QEE_A Sequence Length: 1277 Identity: 82.646% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -3115.031 kcal/mol | |||
| Ramachandra Favored |
Medium |
||||
| QMEANBrane Quality |
High |
||||
| Ramachandran Plot |
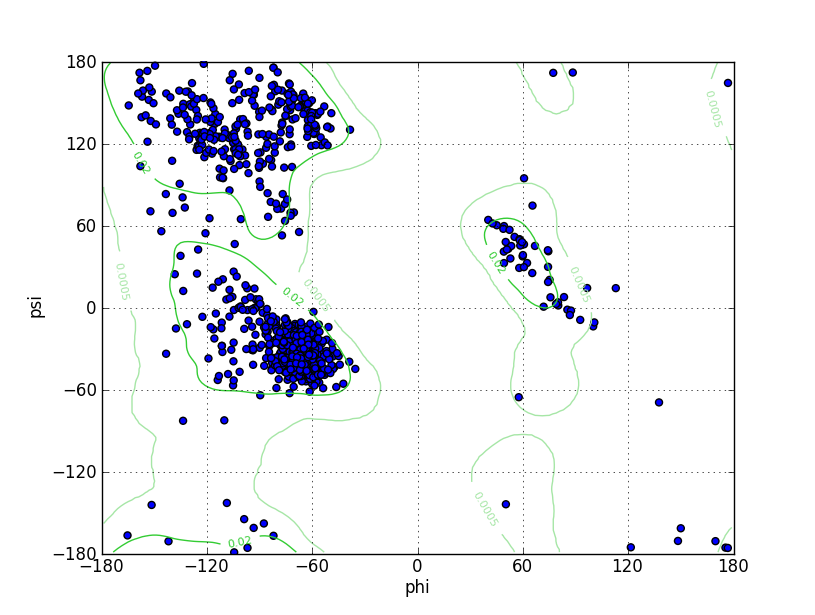
Ramz Z Score: -0.96 ± 0.21 Residues in Favored Region: 1237 Ramachandran favored: 97.02% Number of Outliers: 3 Ramachandran outliers: 0.24% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
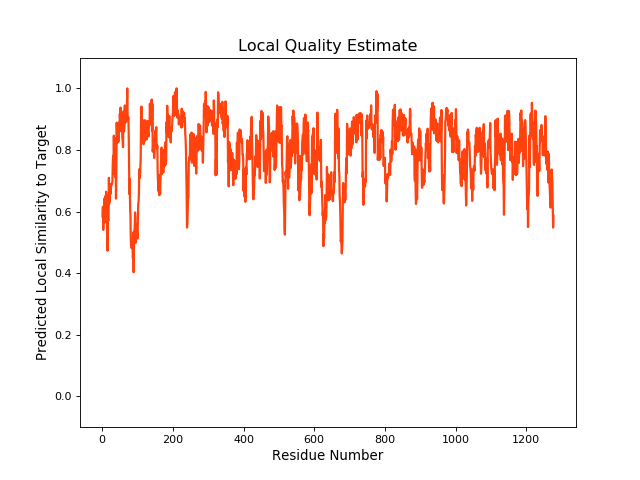
QMEANBrane Score: 0.79 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| References | |||||
| 1 | Structures of the Multidrug Transporter P-glycoprotein Reveal Asymmetric ATP Binding and the Mechanism of Polyspecificity. J Biol Chem. 2017 Jan 13;292(2):446-461. | ||||
| 2 | Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 2009 Mar 27;323(5922):1718-22. | ||||
| 3 | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain. Proc Natl Acad Sci U S A. 2013 Aug 13;110(33):13386-91. | ||||
| 4 | Refined structures of mouse P-glycoprotein. Protein Sci. 2014 Jan;23(1):34-46. | ||||
| 5 | Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein. Acta Crystallogr D Biol Crystallogr. 2015 Mar;71(Pt 3):732-41. | ||||
| 6 | Global marine pollutants inhibit P-glycoprotein: Environmental levels, inhibitory effects, and cocrystal structure. Sci Adv. 2016 Apr 15;2(4):e1600001. | ||||
| 7 | Novel features in the structure of P-glycoprotein (ABCB1) in the post-hydrolytic state as determined at 7.9?? resolution. BMC Struct Biol. 2018 Dec 13;18(1):17. | ||||
| 8 | Structural definition of polyspecific compensatory ligand recognition by P-glycoprotein. IUCrJ. 2020 Jun 6;7(Pt 4):663-672. | ||||
| 9 | RCSB PDB: Structure of Mouse P-Glycoprotein. | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.