Crystal Structure of This DT
Crystallisation Method: EM
Corresponding Chain: A/B/C/D
Sequence Length: 66-1179
Detail: Struture Info
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0512 Transporter Info | ||||
| Gene Name | KCNMA1 | ||||
| Protein Name | Calcium-activated potassium channel subunit alpha-1 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| 3D Structure |
Crystal Structure of This DT Crystallisation Method: EM Corresponding Chain: A/B/C/D Sequence Length: 66-1179 Detail: Struture Info |
||||
| Inter-species Structural Differences (ISD) | |||||
| Canis lupus familiaris (Dog) | |||||
| Gene Name | KCNMA1 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method: Homology modeling Teplate PDB: 6V22_A Sequence Length: 1059 Identity: 97.734% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -2556.092 kcal/mol | |||
| Ramachandra Favored |
Medium |
||||
| QMEANBrane Quality |
Medium |
||||
| Ramachandran Plot |
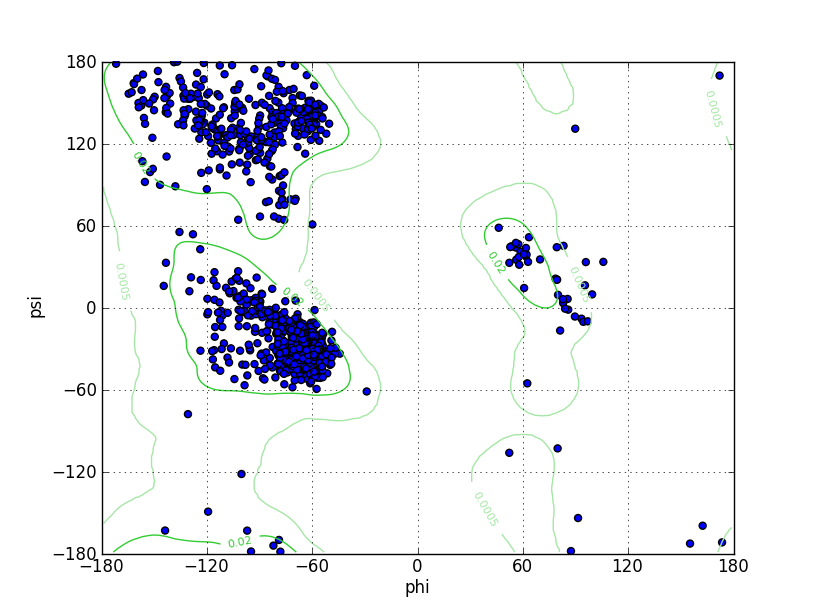
Ramz Z Score: -0.74 ± 0.24 Residues in Favored Region: 1026 Ramachandran favored: 97.07% Number of Outliers: 4 Ramachandran outliers: 0.38% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |

QMEANBrane Score: 0.76 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Inter-species Structural Differences (ISD) | |||||
| Sus scrofa (Pig) | |||||
| Gene Name | KCNMA1 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method: Homology modeling Teplate PDB: 6V22_A Sequence Length: 1059 Identity: 99.433% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -2631.513 kcal/mol | |||
| Ramachandra Favored |
Medium |
||||
| QMEANBrane Quality |
Medium |
||||
| Ramachandran Plot |
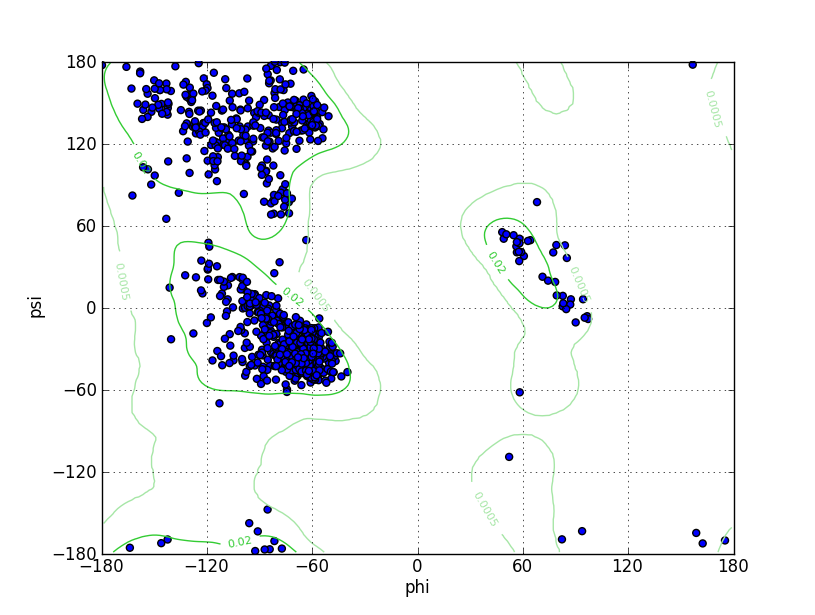
Ramz Z Score: -0.63 ± 0.25 Residues in Favored Region: 1035 Ramachandran favored: 97.92% Number of Outliers: 3 Ramachandran outliers: 0.28% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
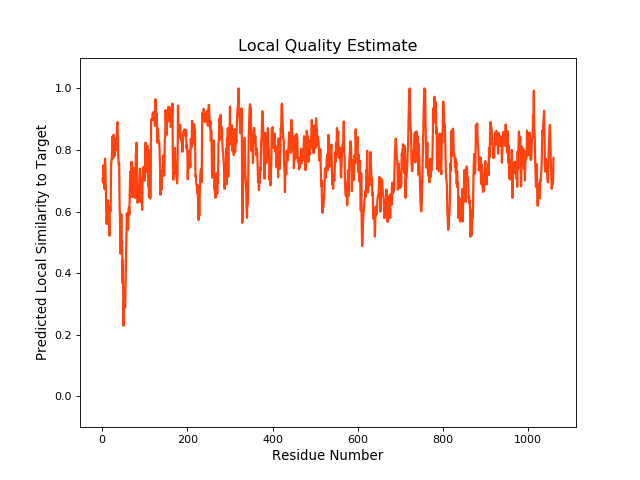
QMEANBrane Score: 0.76 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Inter-species Structural Differences (ISD) | |||||
| Gallus gallus (Chicken) | |||||
| Gene Name | KCNMA1 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method: Homology modeling Teplate PDB: 6V22_A Sequence Length: 1060 Identity: 96.509% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -2543.167 kcal/mol | |||
| Ramachandra Favored |
Medium |
||||
| QMEANBrane Quality |
Medium |
||||
| Ramachandran Plot |
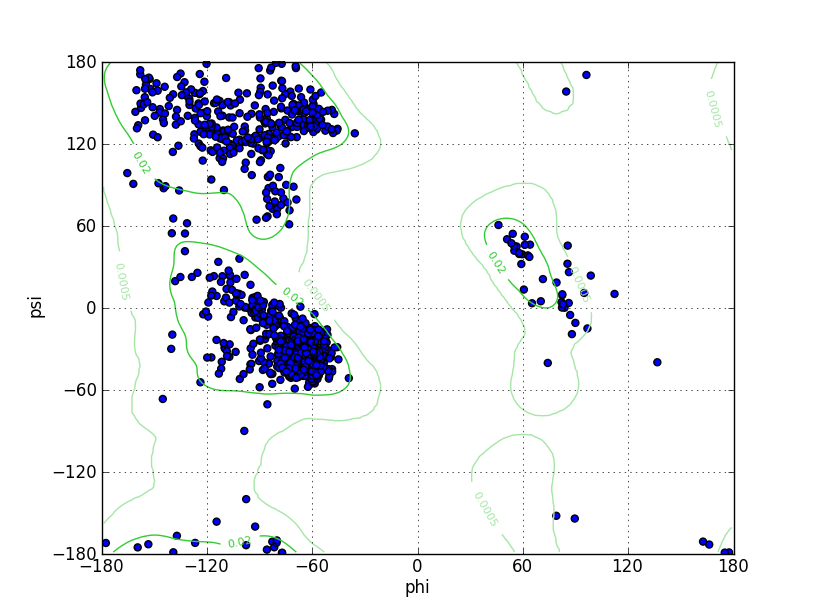
Ramz Z Score: -0.85 ± 0.24 Residues in Favored Region: 1024 Ramachandran favored: 96.79% Number of Outliers: 1 Ramachandran outliers: 0.09% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
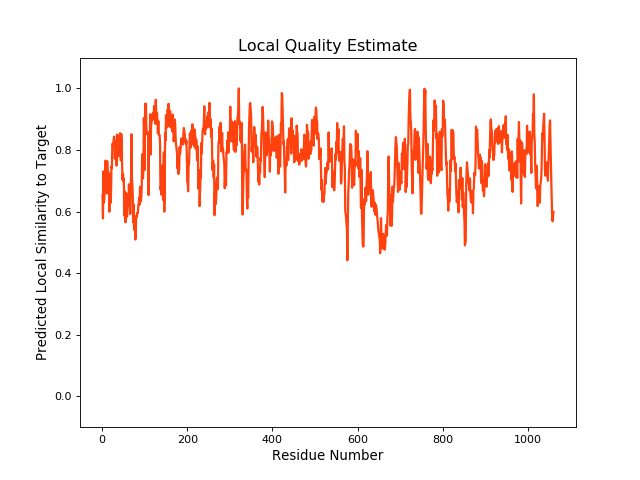
QMEANBrane Score: 0.76 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.