Crystal Structure of This DT
Crystallisation Method: EM
Corresponding Chain: A
Sequence Length: 1-1836
Detail: Struture Info
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0527 Transporter Info | ||||
| Gene Name | SCN4A | ||||
| Protein Name | Voltage-gated sodium channel alpha Nav1.4 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| 3D Structure |
Crystal Structure of This DT Crystallisation Method: EM Corresponding Chain: A Sequence Length: 1-1836 Detail: Struture Info |
||||
| Inter-species Structural Differences (ISD) | |||||
| Equus caballus (Horse) | |||||
| Gene Name | SCN4A | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method: Homology modeling Teplate PDB: 6AGF_A Sequence Length: 1834 Identity: 89.136% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -3106.023 kcal/mol | |||
| Ramachandra Favored |
Medium |
||||
| QMEANBrane Quality |
Medium |
||||
| Ramachandran Plot |
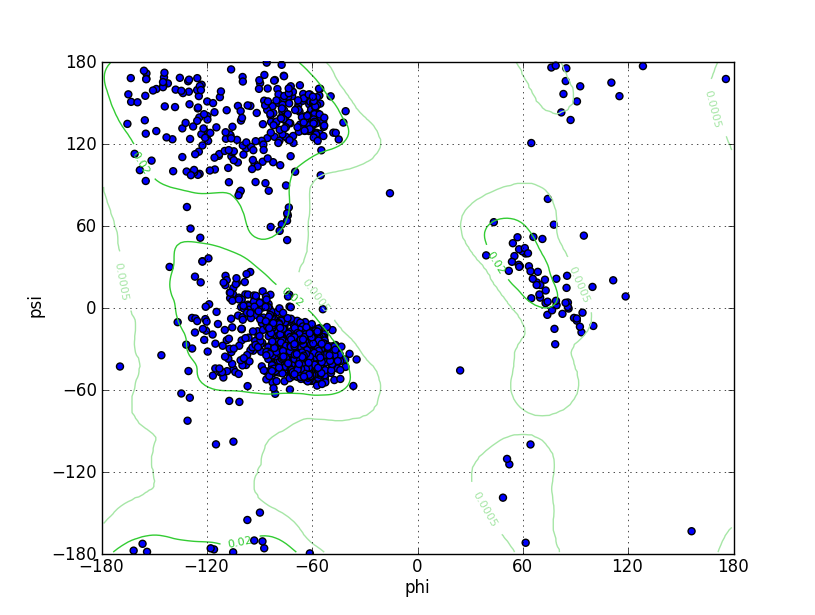
Ramz Z Score: -1.33 ± 0.18 Residues in Favored Region: 1763 Ramachandran favored: 96.23% Number of Outliers: 11 Ramachandran outliers: 0.60% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
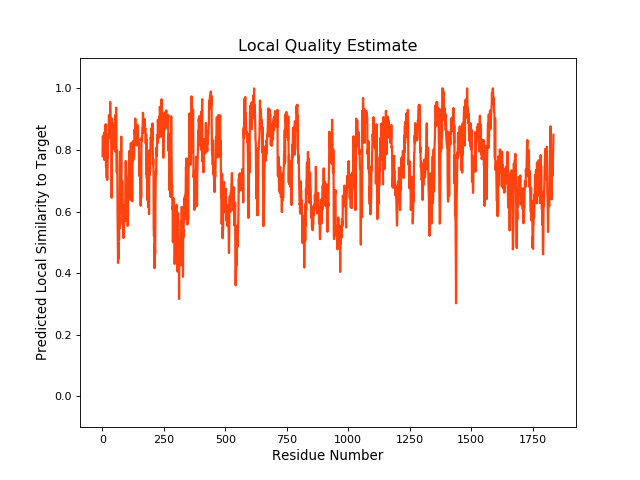
QMEANBrane Score: 0.74 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| References | |||||
| 1 | Crystal structures of Ca(2+)-calmodulin bound to Na(V) C-terminal regions suggest role for EF-hand domain in binding and inactivation. Proc Natl Acad Sci U S A. 2019 May 28;116(22):10763-10772. | ||||
| 2 | RCSB PDB: Veratridine binding to a transmembrane segment of mammalian sodium channel Nav1.4 determined by solid-state NMR. | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.